Machine Learning Infos in AI4(M)S Papers
Update Time: 2025-10-06 12:59:10
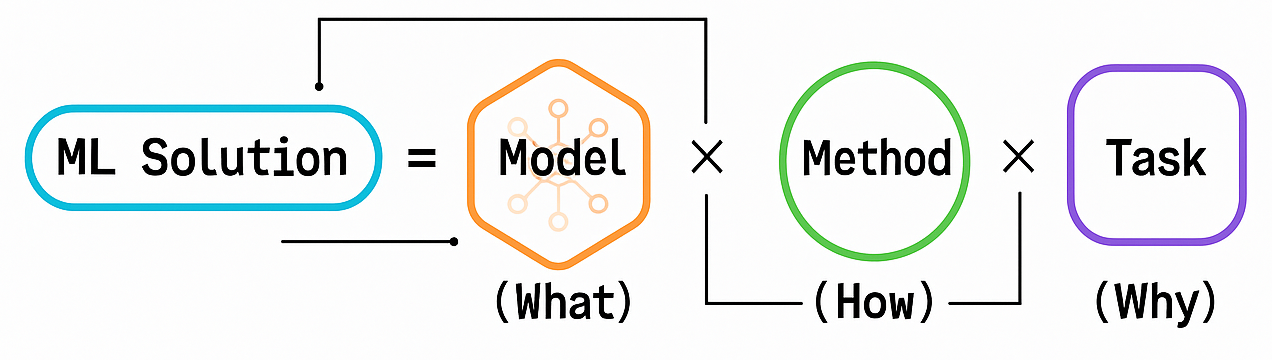
🌳 Machine Learning Taxonomy
🎯 Table 1: Tasks (What to Solve) [16 Categories → 91 Specifics]
| Category | Items |
|---|---|
| Prediction Tasks | Regression, Classification, Binary Classification, Multi-class Classification, Multi-label Classification, Ordinal Regression, Time Series Forecasting, Survival Analysis |
| Ranking and Retrieval | Ranking, Information Retrieval, Recommendation, Collaborative Filtering, Content-Based Filtering |
| Clustering and Grouping | Clustering, Community Detection, Grouping |
| Dimensionality Reduction | Dimensionality Reduction, Feature Selection, Feature Extraction |
| Anomaly and Outlier | Anomaly Detection, Outlier Detection, Novelty Detection, Fraud Detection |
| Density and Distribution | Density Estimation, Distribution Estimation |
| Structured Prediction | Structured Prediction, Sequence Labeling, Named Entity Recognition, Part-of-Speech Tagging, Sequence-to-Sequence |
| Computer Vision Tasks | Image Classification, Object Detection, Object Localization, Semantic Segmentation, Instance Segmentation, Panoptic Segmentation, Pose Estimation, Action Recognition, Video Classification, Optical Flow Estimation, Depth Estimation, Image Super-Resolution, Image Denoising, Image Inpainting, Style Transfer, Image-to-Image Translation, Image Generation, Video Generation |
| Natural Language Processing Tasks | Language Modeling, Text Classification, Sentiment Analysis, Machine Translation, Text Summarization, Question Answering, Reading Comprehension, Dialog Generation, Text Generation, Paraphrase Generation, Text-to-Speech, Speech Recognition, Speech Synthesis |
| Graph Tasks | Node Classification, Link Prediction, Graph Classification, Graph Generation, Graph Matching, Influence Maximization |
| Decision Making | Decision Making, Policy Learning, Control, Planning, Optimization, Resource Allocation |
| Design Tasks | Experimental Design, Hyperparameter Optimization, Architecture Search, AutoML, Neural Architecture Search |
| Association and Pattern | Association Rule Mining, Pattern Recognition, Motif Discovery |
| Matching and Alignment | Entity Matching, Entity Alignment, Record Linkage, Image Matching |
| Generative Tasks | Data Generation, Data Augmentation, Synthetic Data Generation |
| Causal Tasks | Causal Inference, Treatment Effect Estimation, Counterfactual Reasoning |
📊 Table 2: Models (What to Use) [18 Categories → 102 Specifics]
| Category | Items |
|---|---|
| Linear Models | Linear Model, Polynomial Model, Generalized Linear Model |
| Tree-based Models | Decision Tree, Random Forest, Gradient Boosting Tree, XGBoost, LightGBM, CatBoost |
| Kernel-based Models | Support Vector Machine, Gaussian Process, Radial Basis Function Network |
| Probabilistic Models | Naive Bayes, Bayesian Network, Hidden Markov Model, Markov Random Field, Conditional Random Field, Gaussian Mixture Model, Latent Dirichlet Allocation |
| Basic Neural Networks | Perceptron, Multi-Layer Perceptron, Feedforward Neural Network, Radial Basis Function Network |
| Convolutional Neural Networks | Convolutional Neural Network, LeNet, AlexNet, VGG, ResNet, Inception, DenseNet, MobileNet, EfficientNet, SqueezeNet, ResNeXt, SENet, NASNet, U-Net |
| Recurrent Neural Networks | Recurrent Neural Network, Long Short-Term Memory, Gated Recurrent Unit, Bidirectional RNN, Bidirectional LSTM |
| Transformer Architectures | Transformer, BERT, GPT, T5, Vision Transformer, CLIP, DALL-E, Swin Transformer |
| Attention Mechanisms | Attention Mechanism, Self-Attention Network, Multi-Head Attention, Cross-Attention |
| Graph Neural Networks | Graph Neural Network, Graph Convolutional Network, Graph Attention Network, GraphSAGE, Message Passing Neural Network, Graph Isomorphism Network, Temporal Graph Network |
| Generative Models | Autoencoder, Variational Autoencoder, Generative Adversarial Network, Conditional GAN, Deep Convolutional GAN, StyleGAN, CycleGAN, Diffusion Model, Denoising Diffusion Probabilistic Model, Normalizing Flow |
| Energy-based Models | Boltzmann Machine, Restricted Boltzmann Machine, Hopfield Network |
| Memory Networks | Neural Turing Machine, Memory Network, Differentiable Neural Computer |
| Specialized Architectures | Capsule Network, Siamese Network, Triplet Network, Attention Network, Pointer Network, WaveNet, Seq2Seq, Encoder-Decoder |
| Object Detection Models | YOLO, R-CNN, Fast R-CNN, Faster R-CNN, Mask R-CNN, FPN, RetinaNet |
| Time Series Models | ARIMA Model, SARIMA Model, State Space Model, Temporal Convolutional Network, Prophet |
| Pointer Networks | PointNet, PointNet++ |
| Matrix Factorization | Matrix Factorization, Non-negative Matrix Factorization, Singular Value Decomposition |
🎓 Table 3: Learning Methods (How to Learn) [11 Categories → 82 Specifics]
| Category | Items |
|---|---|
| Basic Learning Paradigms | Supervised Learning, Unsupervised Learning, Semi-Supervised Learning, Self-Supervised Learning, Reinforcement Learning |
| Advanced Learning Paradigms | Transfer Learning, Multi-Task Learning, Meta-Learning, Few-Shot Learning, Zero-Shot Learning, One-Shot Learning, Active Learning, Online Learning, Incremental Learning, Continual Learning, Lifelong Learning, Curriculum Learning |
| Training Strategies | Batch Learning, Mini-Batch Learning, Stochastic Learning, End-to-End Learning, Adversarial Training, Contrastive Learning, Knowledge Distillation, Fine-Tuning, Pre-training, Prompt Learning, In-Context Learning |
| Optimization Methods | Gradient Descent, Stochastic Gradient Descent, Backpropagation, Maximum Likelihood Estimation, Maximum A Posteriori, Expectation-Maximization, Variational Inference, Evolutionary Learning |
| Reinforcement Learning Methods | Q-Learning, Policy Gradient, Value Iteration, Policy Iteration, Temporal Difference Learning, Monte Carlo Learning, Actor-Critic, Model-Free Learning, Model-Based Learning, Inverse Reinforcement Learning, Imitation Learning, Multi-Agent Learning |
| Special Learning Settings | Weakly Supervised Learning, Noisy Label Learning, Positive-Unlabeled Learning, Cost-Sensitive Learning, Imbalanced Learning, Multi-Instance Learning, Multi-View Learning, Co-Training, Self-Training, Pseudo-Labeling |
| Domain and Distribution | Domain Adaptation, Domain Generalization, Covariate Shift Adaptation, Out-of-Distribution Learning |
| Collaborative Learning | Federated Learning, Distributed Learning, Collaborative Learning, Privacy-Preserving Learning |
| Ensemble Methods | Ensemble Learning, Bagging, Boosting, Stacking, Blending |
| Representation Learning | Representation Learning, Feature Learning, Metric Learning, Distance Learning, Embedding Learning, Dictionary Learning, Manifold Learning |
| Learning Modes | Generative Learning, Discriminative Learning, Transductive Learning, Inductive Learning |
📈 Summary of Statistics
📑 ML Infos in 361/405 Papers (Chronological Order)
405. A generative artificial intelligence approach for the discovery of antimicrobial peptides against multidrug-resistant bacteria, Nature Microbiology (October 03, 2025)
| Category | Items |
|---|---|
| Datasets | UniProtKB/Swiss-Prot proteome (non-redundant canonical and isoform sequences), AMP dataset (compiled from public AMP databases), Non-AMP dataset (cytoplasm-filtered sequences), External validation dataset (AMPs and non-AMPs), Toxin and non-toxin dataset, Generated sequences from external unconstrained generation models, Generated non-redundant short-peptide datasets (GNRSPDs) from AMPGenix, Non-redundant short-peptide datasets (NRSPDs) constructed from UniProtKB/Swiss-Prot |
| Models | Transformer, GPT, BERT, Random Forest, Support Vector Machine, Multi-Layer Perceptron, Variational Autoencoder, Generative Adversarial Network |
| Tasks | Language Modeling, Text Generation, Binary Classification, Synthetic Data Generation, Data Generation |
| Learning Methods | Self-Supervised Learning, Transfer Learning, Fine-Tuning, Pre-training, Representation Learning |
| Performance Highlights | model_size_parameters: more than 124 million parameters, pretraining_corpus_size: 609,216 protein sequences (Swiss-Prot), generated_sequences_count: 7,798 (AMPGenix default T=1), AUC_benchmarking_set: 0.97, AUPRC_benchmarking_set: 0.96, AUC_test_set: 0.99, AUPRC_test_set: 0.99, Precision: 90.67%, F1_score: 88.89%, MCC: 81.66%, Specificity: 93.93%, Sensitivity: 87.17%, External_validation_precision: 93.99% on independent external validation dataset, AUC_test_set: 0.93, AUPRC_test_set: 0.92, Precision: 84.79%, F1_score: 85.90%, MCC: 72.08%, Specificity: 85.06%, Sensitivity: 87.04%, positive_prediction_rate_on_classifiers: AMPGenix-generated sequences consistently outperformed ProteoGPT across temperature settings when evaluated by 6 AMP classifiers (higher AMP recognition rate), AMPGenix-T1_uniqueness: 0.97, AMPGenix-T1_diversity: 0.98, AMPGenix-T1_novelty: 0.99, AMPGenix-T1_FCD: 10.21, AMPGenix-T2_FCD: 9.26, AMPGenix-T3_FCD: 9.57, ProteoGPT_T1_FCD: 14.87, Macrel_AUC: 0.91 (benchmarking set), Macrel_precision: 95.95% (from Extended Data Table 1) but low sensitivity (52.28%), AmPEP_AUC: not listed; Extended Data Table 1: Precision 32.60%, F1 39.23%, MCC -19.78%, iAMP_Pred_AUC: 0.86 (benchmarking set), iAMP_Pred_precision: 77.86% (Extended Data Table 1), FCD: 13.45 (Extended Data Table 2), uniqueness/diversity/novelty: reported for PepCVAE in Extended Data Table 2 (diversity and novelty ~0.99), FCD: 11.54 (Extended Data Table 2), diversity/novelty: reported in Extended Data Table 2 |
| Application Domains | Antimicrobial peptide (AMP) discovery, Microbiology / infectious disease (multidrug-resistant bacteria: CRAB, MRSA), Protein sequence modeling, Drug discovery / therapeutic peptide design, Computational biology / bioinformatics (sequence mining and generation) |
404. A comprehensive genetic catalog of human double-strand break repair, Science (October 02, 2025)
| Category | Items |
|---|---|
| Datasets | REPAIRome (this study), Toronto KnockOut CRISPR Library v3 (TKOv3), CloneTrackerXP barcode library experiment (representation/depth determination), AAVS1 endogenous locus cut sites (validation), PCAWG tumor cohort (used for mutational signature association), Processed REPAIRome data and code |
| Models | None |
| Tasks | Clustering, Dimensionality Reduction, Feature Selection, Feature Extraction, Information Retrieval, Pattern Recognition |
| Learning Methods | Unsupervised Learning, Representation Learning, Feature Learning |
| Performance Highlights | selected_genes_count: 168, STRING_PPI_enrichment: < 1e-16, GO_enrichment_DSB_repair_FDR: 2.1e-13 (GO:0006302), GO_enrichment_DNA_repair_FDR: 1.3e-11 (GO:0006281), GO_enrichment_NHEJ_FDR: 2.2e-11 (GO:0006303), selection_criteria: distance > 5; FDR < 0.01; replicate PCC > 0.3, UMAP_visualizations: Displayed distance/insertion-deletion/microhomology/editing-efficiency gradients across genes; highlighted NHEJ cluster around LIG4/XRCC4/POLL, examples_distance_values: HLTF distance = 19.7; some genes >10, cosine_similarity_match: VHL knockout effect vector best matched COSMIC indel signature ID11, ID11_prevalence_in_ccRCC: > 50% prevalence (in ccRCC), statistical_tests: prevalence p < 0.001 (Fisher’s exact test); signature activity p < 0.001 (Wilcoxon test); VHL expression (FPKM) associated with active ID11 p < 0.001, correlation_threshold_for_edges: PCC > 0.45, network_genes_count: 183, STRING_PPI_enrichment_of_POLQ_subnetwork: < 1.0e-16, enriched_complexes_in_POLQ_subnetwork: BTRR (FDR = 6.83e-5), Fanconi anemia pathway (FDR = 7.45e-9), SAGA complex (FDR = 3.20e-8) |
| Application Domains | Molecular Biology, Genomics, DNA double-strand break (DSB) repair, CRISPR-Cas gene editing, Cancer Genomics, Computational Biology / Bioinformatics |
402. Machine learning of charges and long-range interactions from energies and forces, Nature Communications (October 01, 2025)
| Category | Items |
|---|---|
| Datasets | Random point-charge gas (this work), KF aqueous solutions (this work), LODE molecular dimer dataset (charged molecular dimers) (ref. 42 / BFDB), SPICE polar dipeptides subset (ref. 43), 4G-HDNNP benchmark datasets (from Ko et al., ref. 12), Pt(111)/KF(aq) dataset (ref. 49), TiO2(101)/NaCl + NaOH + HCl (aq) dataset (ref. 50), LiCl(001)/GaF3(001) interface (this work / generated via on-the-fly FLARE active sampling), Liquid water dataset (ref. 71) used for MD speed benchmarking |
| Models | Multi-Layer Perceptron, Message Passing Neural Network, Graph Neural Network, Ensemble Learning |
| Tasks | Regression, Regression, Feature Extraction, Representation Learning, Anomaly Detection |
| Learning Methods | Supervised Learning, Active Learning, Ensemble Learning, Representation Learning, Backpropagation |
| Performance Highlights | C10H2/C10H3+energy_RMSE_meV_per_atom: 0.73, C10H2/C10H3+_force_RMSE_meV_per_A: 36.9, Na8=9Cl8_energy_RMSE_meV_per_atom: 0.21, Na8=9Cl8_force_RMSE_meV_per_A: 9.78, Au2-MgO(001)_energy_RMSE_meV_per_atom: 0.073, Au2-MgO(001)_force_RMSE_meV_per_A: 7.91, Pt(111)/KF(aq)_energy_RMSE_meV_per_atom: 0.309, Pt(111)/KF(aq)_force_RMSE_meV_per_A: 34.1, TiO2(101)/NaCl+NaOH+HCl(aq)_energy_RMSE_meV_per_atom: 0.435, TiO2(101)/NaCl+NaOH+HCl(aq)_force_RMSE_meV_per_A: 70.5, LiCl/GaF3_ID_force_RMSE_meV_per_A: 78.8, LiCl/GaF3_ID_force_RMSE_meV_per_A_CACE-LR: 67.8, LiCl/GaF3_OOD_force_RMSE_meV_per_A_SR: 116.3, LiCl/GaF3_OOD_force_RMSE_meV_per_A_LR: 40.5, Random_point_charges_charge_prediction_MAE_with_10_configs_e: nearly zero (nearly exact), KF_aq_energy_MAE_meV_per_atom>=100samples: < 0.3, KF_aq_charge_learning_converged_after~couple_hundred_samples: qualitative, Dipole_R2_vs_DFT_on_polar_dipeptides: 0.991, Dipole_MAE_e-angstrom_LE S: 0.089, MBIS_dipole_MAE_e-angstrom: 0.063, Quadrupole_R2_vs_DFT: 0.911, Charges_R2_vs_M BIS: 0.87, Charges_MAE_vs_MBIS_e: 0.24, BEC_diagonal_R2: 0.976, BEC_offdiagonal_R2: 0.838, MD_SR_max_atoms_single_NVIDIA_L40S_GPU: 40000, MD_LR_max_atoms_single_NVIDIA_L40S_GPU: 13000, LR_overhead_vs_SR: minimal with updated implementation (comparable performance) |
| Application Domains | Atomistic simulations of materials, Computational chemistry / molecular modeling, Electrolyte / electrode interfaces (electrochemistry), Ionic solutions and electric double layers, Charged molecular complexes (binding curves), Solid–solid interfaces and heterostructures, Molecular property prediction (dipole, quadrupole, Born effective charges), Machine-learned interatomic potentials (MLIPs) development |
401. Heat-rechargeable computation in DNA logic circuits and neural networks, Nature (October 01, 2025)
| Category | Items |
|---|---|
| Datasets | MNIST (Modified National Institute of Standards and Technology) database, Custom 9-bit two-memory input patterns (L and T patterns), Custom 100-bit two-memory input patterns, Custom Fibonacci-word input patterns (first 16 elements), Synthetic test patterns used for individual circuit component evaluation (e.g., two-input WTA combinations, thresholds) |
| Models | Feedforward Neural Network, Multi-Layer Perceptron |
| Tasks | Binary Classification, Image Classification, Classification, Logic (interpreted as Boolean logic within provided list: Binary Classification / Multi-class Classification not directly but Boolean operations implemented), Sequence-to-Sequence |
| Learning Methods | Unsupervised Learning, Supervised Learning |
| Performance Highlights | reset_success_rate_annihilators_simulated: 93%, reset_success_rate_summation_gates_simulated: 85%, number_of_distinct_strands_in_system: up to 289 distinct strands; 213 present for tested patterns, reusability_rounds_demonstrated: 10 rounds of sequential tests (experiments) with consistent performance; simulations and experiments closely matched, time_to_reset: heating to 95°C and cooling to 20°C in 1 min (reset protocol), rounds_of_computation: 16 rounds (all possible 4-bit inputs), resets_demonstrated: 15 resets over 640 hours, consistency: maintained consistent performance across 16 rounds, kinetics_difference_before_fix: >10-fold difference in kinetics between two hairpin gates sharing same toehold but differing in long domains, reset_success_rates_for_pair_designs: simulations applied 90% and 86% reset success rates for two gates to explain experiments (Extended Data Fig. 6/7), correct_computation_combinations_tested: six input combinations tested with correct behaviour and preserved after reset, reaction_completion_with_hairpin_downstream: approx. 60% reaction completion at high input concentration, reaction_completion_with_two-stranded_downstream: restored full reaction completion, signal_amplification: 10-fold signal amplification within 2 h for chosen catalyst design, rounds_demonstrated: 10 rounds, reusability: consistent off state over 10 rounds when unique inhibitors used, sensitivity_to_inhibitor_quality: performance decay with universal inhibitor due to 5% effective concentration deviation and increasing leak |
| Application Domains | DNA nanotechnology / molecular programming, Synthetic biology, Molecular computing, Molecular diagnostics (potential application), Programmable molecular machines / autonomous chemical systems, Origin-of-life / prebiotic chemistry (conceptual inspiration for heat stations) |
400. Predictive model for the discovery of sinter-resistant supports for metallic nanoparticle catalysts by interpretable machine learning, Nature Catalysis (September 29, 2025)
| Category | Items |
|---|---|
| Datasets | NN-MD generated Pt NP / metal–oxide support dataset (203 systems), OC22-derived candidate surface set (10,662 DFT-relaxed unary and binary metal–oxide surfaces), iGAM training/test split (177 train / 26 test), Experimental adhesion reference: Pt/MgO(100), Experimental sintering test dataset (Pt NPs on α-Al2O3, CeO2, BaO) |
| Models | Generalized Linear Model, Feedforward Neural Network, Ensemble Learning |
| Tasks | Regression, Data Generation, Feature Selection, Ranking, Feature Extraction |
| Learning Methods | Supervised Learning, Active Learning, Pre-training, Ensemble Learning, Transfer Learning |
| Performance Highlights | MAE(Eadh)_train: 0.15 J m^-2, R2_train: 0.90, MAE(Eadh)_test: 0.19 J m^-2, R2_test: 0.79, MAE(Eadh)_6feature: 0.29 J m^-2, MAE(theta)_6feature: ≈10°, simulated_systems: 203, MD_length_each: 500 ps (5×10^5 time steps of 1 fs), temperature: 800 °C, top4_feature_importance_fraction: >80% |
| Application Domains | heterogeneous catalysis, supported metal nanoparticle catalysts (Pt on metal-oxide supports), materials discovery / high-throughput catalyst screening, computational materials science (DFT + ML + MD), nanocatalyst stability / sintering resistance |
399. InterPLM: discovering interpretable features in protein language models via sparse autoencoders, Nature Methods (September 29, 2025)
| Category | Items |
|---|---|
| Datasets | UniRef50 (5 million random protein sequences), Swiss-Prot (UniprotKB reviewed subset; sampled 50,000 proteins), AlphaFold Database (AFDB-F1-v4), InterPro annotations (used for validation of missing annotations), ESM-2 embeddings (pretrained model outputs) |
| Models | Transformer, Autoencoder, Hidden Markov Model |
| Tasks | Feature Extraction, Clustering, Binary Classification, Dimensionality Reduction, Language Modeling, Text Generation, Regression, Feature Selection, Clustering (feature activation patterns: structural vs sequential) |
| Learning Methods | Self-Supervised Learning, Unsupervised Learning, Pre-training, Transfer Learning, Representation Learning, Supervised Learning |
| Performance Highlights | max_features_with_strong_concept_alignment_in_layer: 2,309 (ESM-2-8M layer 5), features_identified_by_SAE_vs_neurons: SAEs extract 3× the concepts found in 8M ESM neurons and 7× in 650M ESM neurons (summary), expansion_factors: 32× (320→10,240 features for ESM-2-8M), 8× (1,280→10,240 for ESM-2-650M), SAE_feature_max_F1_range_on_ESM-2-650M: 0.95–1.0 (maximum F1 scores observed for SAE features), neuron_max_F1_range: 0.6–0.7 (neurons), concepts_detected_ESM-2-650M_vs_ESM-2-8M: 427 vs 143 concepts (≈1.7× more concepts in 650M subset), example_feature_specificity_f1503_F1: 0.998, other_TBDR_cluster_feature_F1s: 0.793, 0.611, glycine_feature_F1s: 0.995, 0.990, 0.86 (highly glycine-specific features), steering_effects: Steering periodic glycine features increased predicted probability of glycine at both steered and masked positions; effect propagated to multiple subsequent periodic repeats with diminishing intensity (quantitative probability changes shown in Fig. 6; steer amounts up to 2.5× maximum activation), median_Pearson_r_for_LLM_generated_descriptions: 0.72 (median across 1,240 features), example_feature_correlations: 0.83, 0.73, 0.99 (example features shown in Fig. 4), example_confirmation: Independent confirmation of Nudix motif in B2GFH1 via HMM-based InterPro annotation (qualitative validation) |
| Application Domains | protein modeling, protein engineering, computational biology, bioinformatics (protein annotation), model interpretability / mechanistic interpretability, biological discovery (novel motif/domain identification) |
398. SimpleFold: Folding Proteins is Simpler than You Think, Preprint (September 27, 2025)
| Category | Items |
|---|---|
| Datasets | Protein Data Bank (PDB), AFDB SwissProt subset (from AlphaFold Protein Structure Database), AFESM (representative clusters), AFESM-E (extended AFESM), CAMEO22 benchmark, CASP14 benchmark (subset), ATLAS (MD ensemble dataset) |
| Models | Transformer, Attention Mechanism, Multi-Head Attention, Pretrained Transformer (ESM2-3B embeddings) |
| Tasks | Regression, Sequence-to-Sequence, Synthetic Data Generation, Distribution Estimation, Multi-class Classification |
| Learning Methods | Generative Learning, Pre-training, Fine-Tuning, Transfer Learning, Supervised Learning, Representation Learning |
| Performance Highlights | CAMEO22TM-score_mean/median: 0.837 / 0.916, CAMEO22_GDT-TS_mean/median: 0.802 / 0.867, CAMEO22_LDDT_mean/median: 0.773 / 0.802, CAMEO22_LDDT-Cα_mean/median: 0.852 / 0.884, CAMEO22_RMSD_mean/median: 4.225 / 2.175, CASP14_TM-score_mean/median: 0.720 / 0.792, CASP14_GDT-TS_mean/median: 0.639 / 0.703, CASP14_LDDT_mean/median: 0.666 / 0.709, CASP14_LDDT-Cα_mean/median: 0.747 / 0.829, CASP14_RMSD_mean/median: 7.732 / 3.923, SimpleFold-100M_CAMEO22_TM-score_mean/median: 0.803 / 0.878, SimpleFold-360M_CAMEO22_TM-score_mean/median: 0.826 / 0.905, SimpleFold-700M_CAMEO22_TM-score_mean/median: 0.829 / 0.915, SimpleFold-1.1B_CAMEO22_TM-score_mean/median: 0.833 / 0.924, SimpleFold-1.6B_CAMEO22_TM-score_mean/median: 0.835 / 0.916, SimpleFold-100M_CASP14_TM-score_mean/median: 0.611 / 0.628, SimpleFold-360M_CASP14_TM-score_mean/median: 0.674 / 0.758, SimpleFold-700M_CASP14_TM-score_mean/median: 0.680 / 0.767, SimpleFold-1.1B_CASP14_TM-score_mean/median: 0.697 / 0.796, SimpleFold-1.6B_CASP14_TM-score_mean/median: 0.712 / 0.801, Pairwise_RMSD_r_no_tuning: 0.44, Global_RMSF_r_no_tuning: 0.45, Per_target_RMSF_r_no_tuning: 0.60, RMWD_no_tuning: 4.22, MD_PCA_W2_no_tuning: 1.62, Joint_PCA_W2_no_tuning: 2.59, %PC_sim>0.5no_tuning: 28, Weak_contacts_J_no_tuning: 0.36, Transient_contacts_J_no_tuning: 0.27, Exposed_residue_J_no_tuning: 0.39, Exposed_MI_matrix_rho_no_tuning: 0.24, Pairwise_RMSD_r_tuned_SF-MD-3B: 0.45, Global_RMSF_r_tuned_SF-MD-3B: 0.48, Per_target_RMSF_r_tuned_SF-MD-3B: 0.67, RMWD_tuned_SF-MD-3B: 4.17, MD_PCA_W2_tuned_SF-MD-3B: 1.34, Joint_PCA_W2_tuned_SF-MD-3B: 2.18, %PC_sim>0.5_tuned_SF-MD-3B: 38, Weak_contacts_J_tuned_SF-MD-3B: 0.56, Transient_contacts_J_tuned_SF-MD-3B: 0.34, Exposed_residue_J_tuned_SF-MD-3B: 0.60, Exposed_MI_matrix_rho_tuned_SF-MD-3B: 0.32, SimpleFold-3B_Apo/holo_res_flex_global: 0.639, SimpleFold-3B_Apo/holo_res_flex_per-target_mean/median: 0.550 / 0.552, SimpleFold-3B_Apo/holo_TM-ens_mean/median: 0.893 / 0.916, SimpleFold-3B_Fold-switch_res_flex_global: 0.292, SimpleFold-3B_Fold-switch_res_flex_per-target_mean/median: 0.288 / 0.263, SimpleFold-3B_Fold-switch_TM-ens_mean/median: 0.734 / 0.766, pLDDT_vs_LDDT-Cα_Pearson_correlation: 0.77, SimpleFold-3B_inference_time_200steps_seq256_s: 15.6, SimpleFold-3B_inference_time_200steps_seq512_s: 27.8, SimpleFold-3B_inference_time_500steps_seq256_s: 37.2, SimpleFold-100M_inference_time_200steps_seq256_s: 3.8 |
| Application Domains | Protein structure prediction / computational structural biology, Molecular dynamics ensemble generation / protein flexibility modeling, De novo protein design / protein generation, Drug discovery (ensemble observables and cryptic pocket identification), Generative modeling for scientific domains (analogy to text-to-image / text-to-3D) |
397. Design of a potent interleukin-21 mimic for cancer immunotherapy, Science Immunology (September 26, 2025)
| Category | Items |
|---|---|
| Datasets | PDB structures (hIL-21/hIL-21R: PDB 3TGX; native hIL-21 complex PDB 8ENT; hγc complex PDB 7S2R; 21h10 complex PDB 9E2T), Computational design candidate set (Rosetta-generated designs), MC38 syngeneic murine tumor model (MC38 adenocarcinoma), B16F10 murine melanoma model (with adoptive TRP1high/low T cell transfer), LCMV-infected mice (virus-specific CD8 T cell analysis), PDOTS (patient-derived organotypic tumor spheroids) from advanced melanoma patients, Bulk RNA-seq (murine CD8 T cells treated in vitro), Single-cell RNA-seq (scRNA-seq) of tumor-infiltrating CD45+ cells, Crystallography diffraction data (21h10/hIL-21R/hγc complex) |
| Models | Message Passing Neural Network, Graph Neural Network, Other (non-ML computational tools) |
| Tasks | Synthetic Data Generation, Clustering, Dimensionality Reduction, Feature Extraction, Data Generation, Image Classification |
| Learning Methods | Supervised Learning, Generative Learning, Unsupervised Learning |
| Performance Highlights | functional_outcome: Generated variant 21AT36 binds IL-21R but not γc; antagonist did not induce STAT phosphorylation, context_readouts: 21AT36 did not induce STAT phosphorylation in murine CD8 T cells; in MC38 tumors an equimolar dose of 21AT36 did not show antitumor activity (Fig. 2F and 2G), design_pool_size: 185 Rosetta designs generated and filtered; downstream selection and mutagenesis led to 21h10, structural_validation: Crystal structure of 21h10 complex solved (PDB ID: 9E2T) with resolution between 2.3 and 3.4 Å, single_cell_input: ≈4000 cells per tumor; 10 mice (PBS, 21h10) or 5 mice (Neo-2/15, mIL-21) per group pooled; 50,000 read pairs per cell sequencing depth, biological_findings: Identification of multiple immune and nonimmune clusters; 21h10 expanded highly activated CD8 T cells and TRP1low tumor-specific T cells and decreased Treg frequency, design_to-function: 21h10 showed STAT1/STAT3 phosphorylation potency equivalent to native hIL-21 and mIL-21 in both human and murine cells; 21h10 elicits similar gene-expression profile at 100 pM compared with 1 nM mIL-21, thermal_stability: 21h10 melting temperature (Tm) ≈ 75°C |
| Application Domains | cancer immunotherapy, computational protein design / de novo protein design, structural biology (X-ray crystallography), immunology (T cell biology, cytokine signaling), single-cell transcriptomics / tumor microenvironment profiling, ex vivo functional profiling of human tumors (PDOTS) |
396. EpiAgent: foundation model for single-cell epigenomics, Nature Methods (September 25, 2025)
| Category | Items |
|---|---|
| Datasets | Human-scATAC-Corpus, Buenrostro2018, Kanemaru2023, Li2023b, Ameen2022, Li2023a, Lee2023, Zhang2021, Pierce2021, Liscovitch-Brauer2021, Long et al. (ccRCC single-cell multi-omics), 10x Genomics single-cell multi-omics human brain dataset |
| Models | Transformer, Attention Mechanism, Multi-Head Attention, Graph Neural Network, Multi-Layer Perceptron, Feedforward Neural Network, Convolutional Neural Network, Support Vector Machine, Transformer, Graph Neural Network |
| Tasks | Feature Extraction, Dimensionality Reduction, Multi-class Classification, Data Generation, Treatment Effect Estimation, Counterfactual Reasoning, Domain Adaptation, Zero-Shot Learning, Clustering |
| Learning Methods | Pre-training, Fine-Tuning, Self-Supervised Learning, Supervised Learning, Transfer Learning, Zero-Shot Learning, Domain Adaptation, Representation Learning, Graph Neural Network |
| Performance Highlights | NMI: higher than all six baseline methods after fine-tuning (exact numeric values reported in Supplementary materials), ARI: higher than all six baseline methods after fine-tuning (exact numeric values reported in Supplementary materials), accuracy_improvement_vs_second_best: 11.036% (average), macro_F1_improvement_vs_second_best: 21.549% (average), NMI_improvement_over_raw: 11.123% (average), ARI_improvement_over_raw: 18.605% (average), Pearson_correlation_median: >0.8 (between imputed signals and average raw signals of corresponding cell types), R2_top_1000_DA_cCREs: >0.7, direction_accuracy_top_100_DA_cCREs: >90% (average across cell types), Wasserstein_distance: lower than baselines (better alignment of predicted and real perturbed cell distributions), Pearson_correlation_vs_GEARS_on_Pierce2021: EpiAgent Pearson correlation average 24.177% higher than GEARS, direction_accuracy_top_DA_cCREs: EpiAgent significantly higher than GEARS (GEARS near random), NMI/ARI/kBET/iLISI: EpiAgent achieves best overall performance vs baselines on clustering and batch-correction metrics (exact numeric values in Fig.4e and Supplementary Figs.), embedding_quality: Zero-shot EpiAgent competitive with baselines on datasets with similar cell populations to pretraining corpus (notably Li2023b), clustering_and_metrics: visual and metric superiority reported (UMAP separation; NMI/ARI comparisons in Fig.2b,c), EpiAgent-B_accuracy: >0.88, EpiAgent-NT_accuracy: >0.95, balanced_accuracy_and_macro_F1: >0.8 (for per-cell-type performance), Wasserstein_distance_change_significance: one-sided t-tests yield P values < 0.05 for majority of perturbations (Liscovitch-Brauer2021 dataset), directional_shift_in_synthetic_ccRCC_experiment: knockouts (EGLN3, ABCC1, VEGFA, Group) shift synthetic cells away from cancer-proportion profiles (quantified by average change in proportion of cancer cell-derived cCREs; exact numeric values in Fig.5g) |
| Application Domains | single-cell epigenomics (scATAC-seq), cell type annotation and atlas construction (human brain, normal tissues), hematopoiesis and developmental trajectory analysis, cardiac niches and heart tissues, stem cell differentiation, cancer epigenetics (clear cell renal cell carcinoma), perturbation response prediction (drug stimulation, CRISPR knockouts), batch correction and multi-dataset integration |
395. Activation entropy of dislocation glide in body-centered cubic metals from atomistic simulations, Nature Communications (September 24, 2025)
| Category | Items |
|---|---|
| Datasets | Fe and W MLIP training datasets (extended from refs. 23, 26), PAFI sampling datasets (finite-temperature sampled configurations along reaction coordinates), Empirical potential (EAM) reference calculations, Experimental yield stress datasets (from literature) |
| Models | Machine-Learning Interatomic Potentials (MLIP), Embedded Atom Method (EAM) potentials |
| Tasks | Regression, Data Generation, Image Classification |
| Learning Methods | Supervised Learning, Transfer Learning |
| Performance Highlights | activation_entropy_harmonic_regime_Fe: ΔS(z2) = 6.3 kB, activation_entropy_difference_above_T0_Fe: ΔS(z2)-ΔS(z1) = 1.6 kB, activation_entropy_harmonic_regime_W: approx. 8 kB, MD_velocity_prefactor_fit_HTST: ν = 3.8×10^9 Hz (HTST fit), MD_velocity_prefactor_fit_VHTST: ν = 9.2×10^10 Hz (variational HTST fit), simulation_cell_size: 96,000 atoms (per atomistic simulation cell), PAFI_computational_cost_per_condition: 5×10^4 to 2.5×10^5 CPU hours (for anharmonic Gibbs energy calculations), Hessian_diagonalization_cost: ≈5×10^4 CPU-hours per atomic system using MLIP, effective_entropy_variation_range_Fe_EAM: ΔSeff varies by ~10 kB between 0 and 700 MPa (Fe, EAM), departure_from_harmonicity_temperature: marked departure from harmonic prediction above ~20 K (Fe, EAM), inverse_Meyer_Neldel_TMNs: Fe: TMN = -406 K (effective fit), W: TMN = -1078 K (effective fit) |
| Application Domains | materials science, computational materials / atomistic simulation, solid mechanics / metallurgy (dislocation glide & yield stress in BCC metals), physics of defects (dislocations, kink-pair nucleation) |
394. Design of facilitated dissociation enables timing of cytokine signalling, Nature (September 24, 2025)
| Category | Items |
|---|---|
| Datasets | PDB structures (design models and solved crystal structures; accession codes 9DCX, 9DCY, 9DCZ, 9DD0, 9DD1, 9DD2, 9DD3, 9DD4, 9DD5, 9OLQ), Designs and analysis code, sequences and source data (Zenodo deposit), Single-molecule tracking (SMT) raw data (Zenodo DOIs: multiple entries), RNA-seq raw data (BioProject PRJNA1302552), SKEMPI database (referenced), Reference genome and gene sets (GRCh38, MSigDB Hallmark gene sets) |
| Models | Diffusion Model |
| Tasks | Clustering, Dimensionality Reduction, Synthetic Data Generation |
| Learning Methods | Unsupervised Learning |
| Performance Highlights | AF2_predicted_RMSD_to_crystal_Cα: <= 1.0 Å, designs_tested_initial_pipeline: 24 designs tested; multiple working designs obtained on first attempt, MD_simulation_length_per_trajectory: 1 μs (triplicate trajectories), agreement_with_DEER_distance_distributions: MD-simulated distance distributions span experimental DEER distribution (qualitative agreement) |
| Application Domains | De novo protein design / computational protein engineering, Structural biology (X-ray crystallography, DEER spectroscopy), Biophysics (kinetic design and measurement of protein–protein interactions), Synthetic biology / biosensor design (rapid luciferase sensors), Immunology / cytokine signalling (design and temporal control of IL-2 mimics), Single-molecule microscopy (live-cell receptor dimerization dynamics), Molecular simulation (MD, integrative modelling) |
393. Tailoring polymer electrolyte solvation for 600 Wh kg−1 lithium batteries, Nature (September 24, 2025)
| Category | Items |
|---|---|
| Datasets | Electrochemical cycling data (coin cells), Electrochemical cycling data (anode-free pouch cells), Materials characterization datasets (NMR, Raman, XPS, TOF-SIMS, TEM, SEM, DSC, GITT, EIS, DEMS), DFT calculation data |
| Models | None |
| Tasks | Experimental Design, Optimization, Data Generation |
| Learning Methods | None |
| Performance Highlights | None |
| Application Domains | Battery materials, Solid-state batteries, Lithium metal batteries (Li-rich Mn-based layered oxide cathodes), Electrochemistry, Energy storage, Materials science / polymer electrolytes, Computational chemistry (DFT) |
392. EDBench: Large-Scale Electron Density Data for Molecular Modeling, Preprint (September 24, 2025)
| Category | Items |
|---|---|
| Datasets | EDBench, ED5-EC, ED5-OE, ED5-MM, ED5-OCS, ED5-MER, ED5-EDP, PCQM4Mv2, Referenced QC datasets (QM7, QM9, QM7-X, PubChemQC, MD17, MD22, WS22, QH9, MultiXC-QM9, MP, ECD, QMugs, ∇2DFT, QM9-VASP, Materials Project) |
| Models | Transformer, Multi-Layer Perceptron, Graph Neural Network |
| Tasks | Regression, Binary Classification, Information Retrieval, Density Estimation, Data Generation |
| Learning Methods | Supervised Learning, Contrastive Learning, Pre-training, Fine-Tuning, Representation Learning, End-to-End Learning |
| Performance Highlights | E1_MAE: 243.49 ± 74.72, E2_MAE: 325.65 ± 160.17, E3_MAE: 858.77 ± 496.74, E4_MAE: 389.24 ± 217.51, E5_MAE: 17.54 ± 10.85, E6_MAE: 243.49 ± 74.73, E1_MAE: 190.77 ± 1.98, E2_MAE: 109.21 ± 2.82, E3_MAE: 369.88 ± 1.34, E4_MAE: 150.05 ± 0.27, E5_MAE: 8.13 ± 0.51, E6_MAE: 190.77 ± 1.98, HOMO-2_MAE_x100: 1.73 ± 0.01, HOMO-1_MAE_x100: 1.68 ± 0.01, HOMO-0_MAE_x100: 1.92 ± 0.01, LUMO+0_MAE_x100: 3.08 ± 0.05, LUMO+1_MAE_x100: 2.86 ± 0.05, LUMO+2_MAE_x100: 3.05 ± 0.02, LUMO+3_MAE_x100: 3.01 ± 0.02, HOMO-2_MAE_x100: 1.75 ± 0.02, HOMO-1_MAE_x100: 1.72 ± 0.02, HOMO-0_MAE_x100: 1.98 ± 0.00, LUMO+0_MAE_x100: 3.21 ± 0.01, LUMO+1_MAE_x100: 3.02 ± 0.02, LUMO+2_MAE_x100: 3.25 ± 0.04, LUMO+3_MAE_x100: 3.20 ± 0.03, Dipole_X_MAE: 0.9123 ± 0.0203, Dipole_Y_MAE: 0.9605 ± 0.0053, Dipole_Z_MAE: 0.7540 ± 0.0068, Magnitude_MAE: 0.7397 ± 0.0467, Dipole_X_MAE: 0.8818 ± 0.0010, Dipole_Y_MAE: 0.9427 ± 0.0008, Dipole_Z_MAE: 0.7416 ± 0.0023, Magnitude_MAE: 0.6820 ± 0.0005, Accuracy: 55.57 ± 2.14, ROC-AUC: 55.97 ± 5.17, AUPR: 57.62 ± 3.91, F1-Score: 66.96 ± 2.08, Accuracy: 57.65 ± 0.18, ROC-AUC: 60.48 ± 0.38, AUPR: 61.54 ± 0.31, F1-Score: 61.41 ± 1.02, GeoFormer + PointVector_ED→MS_Top-1: 17.67 ± 2.10, GeoFormer + PointVector_ED→MS_Top-3: 46.09 ± 4.53, GeoFormer + PointVector_ED→MS_Top-5: 67.63 ± 5.92, GeoFormer + PointVector_MS→ED_Top-1: 27.01 ± 1.69, GeoFormer + PointVector_MS→ED_Top-3: 59.02 ± 2.49, GeoFormer + PointVector_MS→ED_Top-5: 77.42 ± 3.01, GeoFormer + X-3D_ED→MS_Top-1: 68.32 ± 3.70, GeoFormer + X-3D_ED→MS_Top-3: 92.18 ± 2.41, GeoFormer + X-3D_ED→MS_Top-5: 97.31 ± 1.29, GeoFormer + X-3D_MS→ED_Top-1: 70.01 ± 2.93, GeoFormer + X-3D_MS→ED_Top-3: 92.08 ± 2.01, GeoFormer + X-3D_MS→ED_Top-5: 97.17 ± 0.92, EquiformerV2 + PointVector_ED→MS_Top-1: 10.24 ± 1.28, EquiformerV2 + PointVector_ED→MS_Top-3: 32.47 ± 2.69, EquiformerV2 + PointVector_ED→MS_Top-5: 53.42 ± 2.67, EquiformerV2 + PointVector_MS→ED_Top-1: 22.18 ± 0.64, EquiformerV2 + PointVector_MS→ED_Top-3: 54.61 ± 2.89, EquiformerV2 + PointVector_MS→ED_Top-5: 76.83 ± 2.90, EquiformerV2 + X-3D_ED→MS_Top-1: 78.71 ± 0.69, EquiformerV2 + X-3D_ED→MS_Top-3: 94.78 ± 0.40, EquiformerV2 + X-3D_ED→MS_Top-5: 98.13 ± 0.07, EquiformerV2 + X-3D_MS→ED_Top-1: 78.36 ± 0.65, EquiformerV2 + X-3D_MS→ED_Top-3: 94.19 ± 0.14, EquiformerV2 + X-3D_MS→ED_Top-5: 97.74 ± 0.29, ρτ=0.1_MAE: 0.3362 ± 0.2900, ρτ=0.1_Pearson(%): 81.0 ± 8.1, ρτ=0.1_Spearman(%): 56.4 ± 13.7, ρτ=0.1_Time_sec_per_mol: 0.024, ρτ=0.15_MAE: 0.0463 ± 0.0157, ρτ=0.15_Pearson(%): 98.0 ± 6.3, ρτ=0.15_Spearman(%): 87.0 ± 2.7, ρτ=0.15_Time_sec_per_mol: 0.015, ρτ=0.2_MAE: 0.0448 ± 0.0133, ρτ=0.2_Pearson(%): 99.2 ± 0.8, ρτ=0.2_Spearman(%): 91.0 ± 9.1, ρτ=0.2_Time_sec_per_mol: 0.013, DFT_Time_sec_per_mol_for_comparison: 245.8, ED5-EDP_MAE: 0.018 ± 0.003, ED5-EDP_Pearson: 0.993 ± 0.004, ED5-EDP_Spearman: 0.381 ± 0.162, EDMaterial-EDP_MAE: 0.118 ± 0.029, EDMaterial-EDP_Pearson: 0.918 ± 0.034, EDMaterial-EDP_Spearman: 0.633 ± 0.115, X-3D_original_HOMO-2_MAE_x100: 1.75 ± 0.02, X-3D (full)_HOMO-2_MAE_x100: 1.5797, X-3D_original_HOMO-1_MAE_x100: 1.72 ± 0.02, X-3D (full)_HOMO-1_MAE_x100: 1.6359, X-3D_original_HOMO-0_MAE_x100: 1.98 ± 0.00, X-3D (full)_HOMO-0_MAE_x100: 1.9104, X-3D_original_LUMO+0_MAE_x100: 3.21 ± 0.01, X-3D (full)_LUMO+0_MAE_x100: 2.9981, X-3D_original_LUMO+1_MAE_x100: 3.02 ± 0.02, X-3D (full)_LUMO+1_MAE_x100: 2.7028, X-3D_original_LUMO+2_MAE_x100: 3.25 ± 0.04, X-3D (full)_LUMO+2_MAE_x100: 2.8725, X-3D_original_LUMO+3_MAE_x100: 3.20 ± 0.03, X-3D (full)_LUMO+3_MAE_x100: 2.8708, DFT_E1_MAE: 224.13 ± 43.47, DFT_E2_MAE: 155.85 ± 28.75, DFT_E3_MAE: 451.59 ± 58.53, DFT_E4_MAE: 190.47 ± 25.62, DFT_E5_MAE: 9.57 ± 1.56, DFT_E6_MAE: 224.13 ± 43.47, DFT_Mean: 209.29, HGEGNN(2024)_Mean: 186.38, HGEGNN(2025)_Mean: 196.11, HGEGNN(2026)_Mean: 182.75, ED5-OE_xi=2048_mean_MAE_x100: 2.48, ED5-OE_xi=512_mean_MAE_x100: 2.56, ED5-OE_xi=1024_mean_MAE_x100: 2.75, ED5-OE_xi=4096_mean_MAE_x100: 2.70, ED5-OE_xi=8192_mean_MAE_x100: 2.60 |
| Application Domains | Molecular modeling, Quantum chemistry, Machine-learning force fields (MLFFs), Drug discovery / virtual screening, Materials science (periodic systems / crystalline solids), High-throughput quantum-aware modeling |
391. Active Learning for Machine Learning Driven Molecular Dynamics, Preprint (September 21, 2025)
| Category | Items |
|---|---|
| Datasets | Chignolin protein (in-house benchmark suite) |
| Models | Graph Neural Network |
| Tasks | Regression, Data Generation, Dimensionality Reduction, Distribution Estimation |
| Learning Methods | Active Learning, Supervised Learning |
| Performance Highlights | TICA_W1_before: 1.15023, TICA_W1_after: 0.77003, TICA_W1_percent_change: -33.05%, Bond_length_W1_before: 0.00043, Bond_length_W1_after: 0.00022, Bond_length_W1_percent_change: -48.84%, Bond_angle_W1_before: 0.11036, Bond_angle_W1_after: 0.10148, Bond_angle_W1_percent_change: -8.05%, Dihedral_W1_before: 0.25472, Dihedral_W1_after: 0.36378, Reaction_coordinate_W1_before: 0.15141, Reaction_coordinate_W1_after: 0.38302, loss_function: mean-squared error (MSE) between predicted CG forces and projected AA forces (force matching), W1_TICA_after_active_learning: 0.77003 |
| Application Domains | Molecular Dynamics, Protein conformational modeling, Coarse-grained simulations for biomolecules, ML-driven drug discovery / computational biophysics |
390. 3D multi-omic mapping of whole nondiseased human fallopian tubes at cellular resolution reveals a large incidence of ovarian cancer precursors, Preprint (September 21, 2025)
| Category | Items |
|---|---|
| Datasets | nPOD donor fallopian tube cohort (this paper), Visium Cytassist spatial transcriptomics data (ROIs from donor tubes), CODEX multiplexed imaging dataset (25-marker panel), SRS-HSI spatial metabolomics ROIs, Derived/processed imaging dataset (H&E + IHC stacks) |
| Models | Convolutional Neural Network, Convolutional Neural Network, Clustering (unsupervised) |
| Tasks | Structured Prediction, Object Detection, Clustering, Dimensionality Reduction, Clustering, Dimensionality Reduction, Feature Extraction |
| Learning Methods | Supervised Learning, Transfer Learning, Fine-Tuning, Unsupervised Learning, Dimensionality Reduction |
| Performance Highlights | tissue_segmentation_accuracy: 95.2%, epithelial_subtyping_accuracy: 93.2%, nuclei_segmentations_extracted: 2.19 billion, images_processed_for_nuclei: 2,452 H&E-stained images, auto_highlighted_p53_Ki67_locations: 1,285 (mean 257, median 211 per fallopian tube), pathologist_validated_STICs: 99 STICs identified (13 proliferatively active STICs, 86 proliferative dormant STICs) and 11 p53 signatures across 5 donors, SRS_HSI_PCA_kmeans_cluster_finding: distinct metabolite clusters separating lesion vs control ROIs (qualitative), CODEX_clusters: 30 unsupervised clusters combined into 19 annotated cell phenotypes, single_cell_count: 972,276 cells segmented for CODEX WSI, PAGA_connectivity_insights: identified interactions linking TAMs, regulatory DCs, activated T cells and CD8+ memory T cells; STIC cells associated with proliferating epithelial cells (qualitative topology results) |
| Application Domains | Histopathology / Digital Pathology, Oncology (ovarian cancer precursor detection and characterization), Spatial multi-omics integration (spatial proteomics, spatial transcriptomics, spatial metabolomics), Medical image analysis (3D reconstruction and registration) |
389. De novo Design of All-atom Biomolecular Interactions with RFdiffusion3, Preprint (September 18, 2025)
| Category | Items |
|---|---|
| Datasets | Protein Data Bank (PDB) - all complexes deposited through December 2024, AlphaFold2 (AF2) distillation structures (Hsu et al.), Atomic Motif Enzyme (AME) benchmark, Binder design benchmark targets (PD-L1, InsulinR, IL-7Ra, Tie2, IL-2Ra), DNA binder evaluation targets (PDB IDs: 7RTE, 7N5U, 7M5W), Small-molecule binding benchmark (four molecules: FAD, SAM, IAI, OQO), Experimental enzyme design screening set (esterase / cysteine hydrolase designs), Experimental DNA-binding designs |
| Models | Denoising Diffusion Probabilistic Model, Transformer, U-Net, Attention Mechanism, Cross-Attention |
| Tasks | Synthetic Data Generation, Data Generation, Clustering, Sequence-to-Sequence |
| Learning Methods | Self-Supervised Learning, Pre-training, Fine-Tuning |
| Performance Highlights | inference_speedup_vs_RFD2: approximately 10x, parameters: 168M trainable parameters (RFD3) vs ~350M for AF3, unconditional_refolding_rate: 98% of designs have at least one sequence predicted by AF3 to fold within 1.5 Å RMSD (out of 8 ProteinMPNN sequences), diversity_example: 41 clusters out of 96 generations between length 100-250 (TM-score cutoff 0.5), binder_unique_successful_clusters_RFD3avg: 8.2 (average unique successful clusters per target, TM-score clustering threshold 0.6), binder_unique_successful_clusters_RFD1_avg: 1.4 (comparison), DNA_monomer_pass_rate<5Å_DNA-aligned_RMSD: 8.67%, DNA_dimer_pass_rate_<5Å_DNA-aligned_RMSD: 6.67%, DNA_monomer_pass_rate_interface_fixed_after_LigandMPNN: 6.5%, DNA_dimer_pass_rate_interface_fixed_after_LigandMPNN: 5.5%, small_molecule_binder_success_criteria: AF3: backbone RMSD ≤ 1.5 Å; backbone-aligned ligand RMSD ≤ 5 Å; Interface min PAE ≤ 1.5; ipTM ≥ 0.8, small_molecule_result_summary: RFD3 significantly outperforms RFdiffusionAA across the four tested molecules; RFD3 designs are more diverse, novel relative to training set, and have lower Rosetta ∆∆G binding energies (no single-number aggregates reported in main text), AME_win_count: RFD3 outperforms RFD2 on 37 of 41 cases (90%), AME_residue_islands_>4pass_rate_RFD3: 15%, AME_residue_islands>4_pass_rate_RFD2: 4%, experimental_DNA_binder_screen_results: 5 designs synthesized; 1 bound with EC50 = 5.89 ± 2.15 μM (yeast surface display), experimental_enzyme_screen_results: 190 designs screened; 35 multi-turnover designs observed; best enzyme Kcat/Km = 3557 |
| Application Domains | de novo protein design (generative biomolecular design), protein-protein binder design, protein-DNA binder design, protein-small molecule binder design, enzyme active site scaffolding and enzyme design, symmetric oligomer design, biomolecular modeling and structural prediction (evaluation with AlphaFold3) |
388. DeepSeek-R1 incentivizes reasoning in LLMs through reinforcement learning, Nature (September 17, 2025)
| Category | Items |
|---|---|
| Datasets | AIME 2024, MMLU, MMLU-Redux, MMLU-Pro, DROP, C-Eval, IF-Eval (IFEval), FRAMES, GPQA Diamond, SimpleQA / C-SimpleQA, CLUEWSC, AlpacaEval 2.0, Arena-Hard, SWE-bench Verified, Aider-Polyglot, LiveCodeBench, Codeforces, CNMO 2024 (Chinese National High School Mathematics Olympiad), MATH-500, Cold-start conversational dataset (paper-curated), Preference pairs for helpful reward model, Safety dataset for safety reward model, Released RL prompts and rejection-sampling data samples |
| Models | Transformer, Attention Mechanism |
| Tasks | Question Answering, Reading Comprehension, Multi-class Classification, Text Generation, Sequence-to-Sequence, Information Retrieval |
| Learning Methods | Reinforcement Learning, Supervised Learning, Fine-Tuning, Knowledge Distillation, Policy Gradient |
| Performance Highlights | AIME 2024 (pass@1): 77.9%, AIME 2024 (self-consistency cons@16): 86.7% (with self-consistency decoding), AIME training start baseline (pass@1): 15.6% (initial during RL trajectory), English MMLU (EM): 90.8, MMLU-Redux (EM): 92.9, MMLU-Pro (EM): 84.0, DROP (3-shot F1): 92.2, IF-Eval (Prompt Strict): 83.3, GPQA Diamond (Pass@1): 71.5, SimpleQA (Correct): 30.1, FRAMES (Acc.): 82.5, AlpacaEval 2.0 (LC-winrate): 87.6, Arena-Hard (vs GPT-4-1106): 92.3, Code LiveCodeBench (Pass@1-COT): 65.9, Codeforces (Percentile): 96.3, Codeforces (Rating): 2,029, SWE-bench Verified (Resolved): 49.2, Aider-Polyglot (Acc.): 53.3, AIME 2024 (Pass@1): 79.8, MATH-500 (Pass@1): 97.3, CNMO 2024 (Pass@1): 78.8, CLUEWSC (EM): 92.8, C-Eval (EM): 91.8, C-SimpleQA (Correct): 63.7, Code LiveCodeBench (Pass@1-COT): 63.5, Codeforces (Percentile): 90.5, AIME 2024 (Pass@1): 74.0, MATH-500 (Pass@1): 95.9, CNMO 2024 (Pass@1): 73.9, helpful preference pairs curated: 66,000 pairs, safety annotations curated: 106,000 prompts, qualitative statement: Distilled models “exhibit strong reasoning capabilities, surpassing the performance of their original instruction-tuned counterparts.” |
| Application Domains | Mathematics (math competitions, AIME, CNMO, MATH-500), Computer programming / Software engineering (Codeforces, LiveCodeBench, LiveCodeBench, SWE-bench Verified), Biology (graduate-level problems), Physics (graduate-level problems), Chemistry (graduate-level problems), Instruction following / conversational AI (IF-Eval, AlpacaEval, Arena-Hard), Information retrieval / retrieval-augmented generation (FRAMES), Safety and alignment evaluation (safety datasets, reward models) |
386. Modeling-Making-Modulating High-Entropy Alloy with Activated Water-Dissociation Centers for Superior Electrocatalysis, Journal of the American Chemical Society (September 17, 2025)
| Category | Items |
|---|---|
| Datasets | DFT adsorption dataset for PtPdRhRuMo HEA (this work), Open Catalyst Project pretrained graph neural networks (OC20/OC22), Predicted composition screening set (CatBoost evaluations) |
| Models | CatBoost, Linear Model, Support Vector Machine, Random Forest, Gradient Boosting Tree, XGBoost, Multi-Layer Perceptron, Graph Neural Network, Graph Convolutional Network |
| Tasks | Regression, Hyperparameter Optimization, Feature Selection, Optimization, Representation Learning |
| Learning Methods | Supervised Learning, Pre-training, Transfer Learning, Representation Learning |
| Performance Highlights | MAE_train_eV: ∼0.03, MAE_test_eV: ∼0.07, MAE_test_eV: < 0.1 |
| Application Domains | Electrocatalysis, Methanol Oxidation Reaction (MOR), Catalyst design for energy conversion, High-Entropy Alloy (HEA) materials discovery, Computational materials science (DFT + ML integration) |
385. Learning the natural history of human disease with generative transformers, Nature (September 17, 2025)
| Category | Items |
|---|---|
| Datasets | UK Biobank (first-occurrence disease data), Danish national registries (Danish National Patient Registry, Danish Register of Causes of Death), Delphi-2M-sampled synthetic dataset |
| Models | Transformer, GPT, BERT, Linear Model, Encoder-Decoder |
| Tasks | Language Modeling, Multi-class Classification, Binary Classification, Survival Analysis, Time Series Forecasting, Data Generation, Representation Learning |
| Learning Methods | Self-Supervised Learning, Supervised Learning, Transfer Learning, Stochastic Learning, Backpropagation, Maximum Likelihood Estimation, Representation Learning, End-to-End Learning |
| Performance Highlights | average age–sex-stratified AUC (internal validation, next-token prediction, averaged across diagnoses): ≈0.76, AUC (death, age-stratified, internal validation): 0.97, AUC (long-term, 10 years horizon average): 0.70 (average AUC decreases from ~0.76 to 0.70 after 10 years), calibration: Predicted rates closely match observed counts in calibration analyses in 5-year age brackets (qualitative, shown in Extended Data Fig. 3), time-to-event prediction accuracy (aggregate): Model provides consistent estimates of inter-event times (Fig. 1g and methods describe log-likelihood for exponential waiting times), synthetic-trained-model AUC (age–sex-stratified average on observed validation data): 0.74 (trained exclusively on Delphi-2M synthetic data; ~3 percentage points lower than original Delphi-2M), fraction of correctly predicted disease tokens in year 1 of sampling: 17% (compared with 12–13% using sex and age alone), fraction correct after 20 years: <14%, Dementia AUC (Transformer baseline): 0.79, Death AUC (Transformer baseline): 0.78, CVD AUC (Transformer baseline shown alongside others): 0.69 (Transformer as listed in Fig. 2f) |
| Application Domains | Population-scale human disease progression modeling, Epidemiology / public health planning (disease burden projection), Clinical risk prediction (CVD, dementia, diabetes, death, and >1,000 ICD-10 diagnoses), Synthetic data generation for privacy-preserving biomedical model training, Explainable AI for healthcare (embedding/SHAP-based interpretability), Precision medicine / individualized prognostication |
384. Discovery of Unstable Singularities, Preprint (September 17, 2025)
| Category | Items |
|---|---|
| Datasets | Córdoba-Córdoba-Fontelos (CCF) model collocation data (synthetic, generated in self-similar coordinates), Incompressible Porous Media (IPM) with boundary collocation data (synthetic, generated in self-similar coordinates), 2D Boussinesq (with boundary) collocation data (synthetic, generated in self-similar coordinates) |
| Models | Multi-Layer Perceptron, Feedforward Neural Network |
| Tasks | Regression, Optimization, Hyperparameter Optimization |
| Learning Methods | Self-Supervised Learning, Multi-Stage Training, Backpropagation, Gradient Descent, Stochastic Learning, Mini-Batch Learning |
| Performance Highlights | CCF stable log10(max residual): -13.714, CCF 1st unstable log10(max residual): -13.589, CCF 2nd unstable log10(max residual): -6.664, IPM stable log10(max residual): -11.183, IPM 1st unstable log10(max residual): -10.510, IPM 2nd unstable log10(max residual): -8.101, IPM 3rd unstable log10(max residual): -7.526, Boussinesq stable log10(max residual): -8.178, Boussinesq 1st unstable log10(max residual): -8.038, Boussinesq 2nd unstable log10(max residual): -7.772, Boussinesq 3rd unstable log10(max residual): -7.558, Boussinesq 4th unstable log10(max residual): -7.020, CCF stable residual (order): O(10^-13), CCF 1st unstable residual (order): O(10^-13), IPM stable residual (order): O(10^-11), IPM 1st unstable residual (order): O(10^-10), convergence to O(10^-8) with GN: ≈50k iterations (~3 A100 GPU hours), λ for CCF 1st unstable (from literature and this work agreement): λ1 ≈ 0.6057 (Wang et al.); reproduced/improved here, λ for CCF 2nd unstable (this work): λ2 = 0.4703 (text) |
| Application Domains | Mathematical fluid dynamics, Partial differential equations (PDEs) / numerical analysis, Singularity formation and mathematical physics, Computer-assisted proofs (rigorous numerics) |
382. A Generative Foundation Model for Antibody Design, Preprint (September 16, 2025)
| Category | Items |
|---|---|
| Datasets | SAbDab (training set up to 2022-12-31), SAb23H2 / SAb-23H2-Ab (test set), SAb-23H2-Nano (nanobody test set), PPSM pre-training corpora (UniRef50, PDB multimers, PPI, OAS antibody pairs), IgDesign test set (used for inverse design benchmarking), PD-L1 experimental de novo design dataset (wet-lab candidates), Case-study experimental targets (datasets of antigens used in experiments) |
| Models | Denoising Diffusion Probabilistic Model, Diffusion Model, Transformer, Attention Mechanism, Self-Attention Network, Multi-Head Attention, Graph Neural Network |
| Tasks | Structured Prediction, Sequence-to-Sequence, Data Generation, Optimization, Ranking, Regression |
| Learning Methods | Pre-training, Fine-Tuning, Self-Supervised Learning, Curriculum Learning, Transfer Learning, Representation Learning, Fine-Tuning (task-specific) + Frequency-based selection (screening) |
| Performance Highlights | TM-Score: 0.9591, lDDT: 0.8956, RMSD (antibody): 2.1997 Å, DockQ: 0.2986, iRMS: 6.2195 Å, LRMS: 19.4888 Å, Success Rate (SR, DockQ>0.23): 0.4667, AAR (CDR L2): median ≈ 0.8 (Fig.2f described); (Table B4 AAR values for IgGM: L1 0.750, L2 0.743, L3 0.635, H1 0.740, H2 0.644, H3 0.360), Protein A binding (VHH3 variants): VHH3-4M KD = 387 nM; VHH3-5M KD = 384 nM; VHH3-WT no binding (KD >10,000 nM), Humanization (mouse→human) KD comparisons: Mouse KD = 0.120 nM; Human-1 KD = 0.171 nM; Human-2 KD = 0.195 nM; Human-3 KD = 0.395 nM; Human-4 KD = 0.486 nM; Human-5 KD = 0.139 nM, VH-Humanness Score (average): IgGM designed average 0.909 vs original murine 0.676 (text); CDR3 RMSD: IgGM 0.750 Å vs BioPhi 0.983 Å, Structural preservation backbone RMSD: average backbone RMSD reported as 1.10 ± 0.06 Å, I7 (anti-IL-33) KD improvement: Original KD = 52.02 nM → Best variant KD = 9.753 nM (5.3-fold improvement), I7 EC50 improvements (first round): [‘Mutants M1, M7, M10 achieved 4–6× increase in affinity (ELISA)’], Broadly neutralizing R1-32 variants (SARS-CoV-2 RBD) KD changes: Q61E enabled binding to Lambda and BQ.1.1 from no binding (examples: Q61E-Lambda KD 948 nM; Q61E-BQ.1.1 KD 948 nM), subsequent N58D,Q61E improved BQ.1.1 KD to 107 nM (~9–10× improvement compared to earlier), PD-L1 de novo design success rate: 7/60 candidates had nanomolar or picomolar KD (success rate 7/60 ≈ 11.7%), Top de novo binder KD range: 0.084 nM (D1) to 2.89 nM (D7), Example D1 KD: 0.084 nM; D1 IC50 = 7.29 nM; D1 displaced PD-1 from PD-L1 in competition assays, Ablation impact (w/o PPSM): Comparative ablation: w/o PPSM AAR 0.322 vs full IgGM AAR 0.360; DockQ 0.233 vs 0.246; SR 0.426 vs 0.433 (Table B5), Contextual effect: PPSM improves interface bias and sequence recovery marginally |
| Application Domains | Antibody engineering / design, Structural biology (protein structure prediction and docking), Therapeutic antibody discovery (immuno-oncology PD-L1, anti-viral SARS-CoV-2), Protein engineering (framework optimization, humanization), Computational biology / bioinformatics (protein sequence-structure co-design) |
381. MSnLib: efficient generation of open multi-stage fragmentation mass spectral libraries, Nature Methods (September 15, 2025)
| Category | Items |
|---|---|
| Datasets | MSnLib (this work), MCEBIO library (subset used in analyses), NIH NPAC ACONN (NIHNP), OTAVAPEP (peptidomimetic library), ENAMDISC (Discovery Diverse Set DDS-10 from Enamine), ENAMMOL (Enamine + Molport mixture, incl. carboxylic acid fragment library), MCESCAF (MCE 5K Scaffold Library), MCEDRUG (FDA-approved drugs subset from MCE), Evaluation dataset: drug-incubated bacterial cultures (MSV000096589), Public spectral libraries (comparison references) |
| Models | None |
| Tasks | Classification, Clustering, Dimensionality Reduction, Feature Extraction, Information Retrieval |
| Learning Methods | Supervised Learning, Unsupervised Learning |
| Performance Highlights | None |
| Application Domains | clinical metabolomics, natural product discovery, exposomics, untargeted liquid chromatography–mass spectrometry (LC–MS) annotation, microbial metabolite analysis / metabolomics |
380. Spatial gene expression at single-cell resolution from histology using deep learning with GHIST, Nature Methods (September 15, 2025)
| Category | Items |
|---|---|
| Datasets | BreastCancer1 (10x Xenium), BreastCancer2 (10x Xenium), LungAdenocarcinoma (10x Xenium), Melanoma (10x Xenium), BreastCancerILC and BreastCancerIDC (10x Xenium), HER2ST spatial transcriptomics dataset, NuCLS dataset, TCGA-BRCA (The Cancer Genome Atlas - Breast Invasive Carcinoma), Mixed DCIS cohort (in-house), Single-cell reference datasets (breast, melanoma, lung) |
| Models | U-Net, Convolutional Neural Network, DenseNet, VGG, ResNet, Transformer, Graph Neural Network, Multi-Head Attention, Cross-Attention, Encoder-Decoder, Multi-Layer Perceptron, Linear Model |
| Tasks | Regression, Image-to-Image Translation, Semantic Segmentation, Multi-class Classification, Weakly Supervised Learning, Survival Analysis, Clustering, Feature Extraction |
| Learning Methods | Multitask Learning, Supervised Learning, Weakly Supervised Learning, End-to-End Learning, Pre-training, Self-Supervised Learning, Transfer Learning, Gradient Descent |
| Performance Highlights | cell-type_accuracy_BreastCancer1: 0.75, cell-type_accuracy_BreastCancer2: 0.66, median_PCC_top20_SVGs: 0.7, median_PCC_top50_SVGs: 0.6, gene_corr_SCD: 0.74, gene_corr_FASN: 0.77, gene_corr_FOXA1: 0.8, gene_corr_EPCAM: 0.84, melanoma_celltype_proportion_corr: 0.92, lung_adenocarcinoma_celltype_proportion_corr: 0.97, PCC_all_genes: 0.16, SSIM_all_genes: 0.1, PCC_HVGs: 0.2, PCC_SVGs: 0.27, SSIM_HVGs: 0.17, SSIM_SVGs: 0.26, RMSE_all_genes: 0.2, RMSE_SVGs: 0.22, top_gene_correlations: {‘GNAS’: 0.42, ‘FASN’: 0.42, ‘SCD’: 0.34, ‘MYL12B’: 0.32, ‘CLDN4’: 0.32}, C-index_GHIST: 0.57, C-index_RNASeq_STgene_baseline: 0.55, Kaplan_Meier_logrank_P: 0.017, PCC_all_genes: 0.14, SSIM_all_genes: 0.08, PCC_all_genes: 0.11, SSIM_all_genes: 0.07 |
| Application Domains | Histopathology (H&E imaging), Spatial transcriptomics (subcellular and spot-based SRT), Cancer (breast cancer, HER2+ subtype, luminal cohort), Lung adenocarcinoma, Melanoma, Multi-omics integration (TCGA multi-omics), Biomarker discovery and survival prognosis |
378. Bridging histology and spatial gene expression across scales, Nature Methods (September 15, 2025)
| Category | Items |
|---|---|
| Datasets | 10x Xenium, 10x Visium, The Cancer Genome Atlas (TCGA) H&E slides, Various cancer datasets (breast cancer, lung adenocarcinoma, melanoma), Gastric cancer samples (used with iSCALE), Multiple sclerosis brain tissue (used with iSCALE), Single-cell RNA-sequencing reference atlas, Subcellular-resolution spatial transcriptomics data (general) |
| Models | Multi-Layer Perceptron, Convolutional Neural Network, Encoder-Decoder, Graph Neural Network |
| Tasks | Regression, Multi-Task Learning, Image Super-Resolution, Multi-class Classification, Semantic Segmentation, Synthetic Data Generation |
| Learning Methods | Multi-Task Learning, Supervised Learning, Unsupervised Learning, Transfer Learning |
| Performance Highlights | None |
| Application Domains | Spatial transcriptomics, Histopathology / H&E image analysis, Cancer biology (breast cancer, lung adenocarcinoma, melanoma, gastric cancer), Neuropathology (multiple sclerosis brain tissue), Biobanks and archived clinical cohorts (e.g., TCGA), Digital spatial omics and tissue-wide molecular mapping |
377. Structural Insights into Autophagy in the AlphaFold Era, Journal of Molecular Biology (September 15, 2025)
| Category | Items |
|---|---|
| Datasets | None |
| Models | None |
| Tasks | None |
| Learning Methods | None |
| Performance Highlights | None |
| Application Domains | Autophagy research, Structural biology / protein structure prediction, Molecular biology, Biophysics, Therapeutic/drug discovery (rational drug design) |
376. Scaling up spatial transcriptomics for large-sized tissues: uncovering cellular-level tissue architecture beyond conventional platforms with iSCALE, Nature Methods (September 15, 2025)
| Category | Items |
|---|---|
| Datasets | Gastric cancer Xenium sample - BS06-9313-8_Tumor (Tumor), Gastric Xenium sample - Normal 1 (Gastric Patient 1, Normal 1), Gastric Xenium sample - Normal 2 (Gastric Patient 2, Normal 2), Human large-sized MS brain sample - MS330-AL (MS Sample 1), Human large-sized MS brain sample - MS330-CAL (MS Sample 2), Pseudo-Visium daughter captures (simulated from Xenium full-slide data) |
| Models | Multi-Layer Perceptron, Vision Transformer |
| Tasks | Regression, Semantic Segmentation, Multi-class Classification, Clustering, Feature Extraction, Dimensionality Reduction |
| Learning Methods | Weakly Supervised Learning, Supervised Learning, Transfer Learning, Out-of-Distribution Learning, Mini-Batch Learning, Gradient Descent, End-to-End Learning |
| Performance Highlights | alignment_accuracy: 99% (semiautomatic alignment algorithm accuracy for daughter captures), RMSE: iSCALE-Seq outperformed iStar across RMSE (displayed in Fig. 3a; lower is better), SSIM: iSCALE-Seq outperformed iStar across SSIM (displayed in Fig. 3a; higher is better), Pearson_correlation: iSCALE-Seq achieved higher Pearson correlations vs iStar; ~50% of genes achieved r > 0.45 at 32 µm resolution for iSCALE-Seq; example per-gene r values shown (e.g., iSCALE-Seq r = 0.5037 for one gene in Fig. 3c), adjusted_Rand_index: 0.74 (segmentation result from out-of-sample predictions closely aligns with in-sample segmentation; reported when comparing segmentations in normal gastric out-of-sample experiment), RMSE: iSCALE-Img achieves low RMSE in in-sample evaluations; comparable performance to iSCALE-Seq (Fig. 3a), SSIM: High SSIM relative to competing methods (Fig. 3a), Pearson_correlation: iSCALE-Img had generally low Pearson at superpixel level but improved with larger superpixel sizes; example reported correlations improved with resolution, Spearman_correlation: Out-of-sample predictions: at 64 µm resolution ≈50% of genes achieved Spearman r > 0.45; overall Spearman reported across resolutions (Fig. 4c), chi_squared_concordance: 99 of the top 100 HVGs exhibited significantly concordant out-of-sample predicted expression patterns vs ground truth (chi-squared statistic with Bonferroni correction), adjusted_Rand_index: 0.74 (segmentation agreement between out-of-sample and in-sample segmentations in Normal gastric data) |
| Application Domains | Spatial transcriptomics, Histopathology / digital pathology (H&E image analysis), Oncology (gastric cancer tissue analysis), Neurology / Multiple sclerosis brain tissue analysis, Single-cell and spatial genomics integration |
375. Integrating diverse experimental information to assist protein complex structure prediction by GRASP, Nature Methods (September 15, 2025)
| Category | Items |
|---|---|
| Datasets | PSP dataset, Self-curated benchmark dataset (contact RPR and IR), Simulated XL dataset (SDA XL simulations), Experimental XL dataset (real-world XL cases), CL dataset (covalent labeling), CSP dataset (chemical shift perturbation), Simulated DMS (BM5.5) dataset, Hitawala–Gray simulated DMS dataset, Experimental DMS dataset (SARS-CoV-2 RBD antibodies), Mitochondria in situ XL-MS dataset |
| Models | Transformer, Attention Mechanism, Self-Attention Network, Multi-Head Attention, Ensemble (ensemble prediction from multiple checkpoints) |
| Tasks | Link Prediction, Ranking, Clustering |
| Learning Methods | Fine-Tuning, Pre-training, Transfer Learning, Ensemble Learning, Supervised Learning |
| Performance Highlights | benchmark_mean_DockQ_with_2contact_RPRs: 0.35, benchmark_success_rate>0.23_with_2_contact_RPRs: 52.7%, IRs_mean_DockQ_for_4_10_20_restraints: 0.24 / 0.34 / 0.41, IRs_success_rate_for_4_10_20_restraints: 35.3% / 51.9% / 63.2%, AFM_without_restraints_mean_DockQ: 0.17, AF3_without_restraints_mean_DockQ: 0.23, simulated_XL_mean_DockQ_1%: 0.18, simulated_XL_mean_DockQ_2%: 0.21, simulated_XL_mean_DockQ_5%: 0.27, HADDOCK_mean_DockQ_1%_2%_5%: 0.06 / 0.08 / 0.10, AlphaLink_mean_DockQ_1%_2%_5%: 0.12 / 0.13 / 0.20, restraint_satisfaction_median_at_2%_coverage_all_correct: 69% (all) / 81% (correct restraints), iterative_noise_filtering_effect: improved DockQ and pLDDT across coverage levels, experimental_XL_mean_DockQ_for_9_samples: 0.48 (GRASP) vs 0.31 (AFM) vs 0.38 (AlphaLink) vs 0.05 (ClusPro) vs 0.05 (HADDOCK), example_4G3Y_DockQ: GRASP 0.77 vs AFM 0.03; AlphaLink 0.67; HADDOCK 0.02, CL_average_DockQ_GRASP: 0.58, CL_average_DockQ_AFM: 0.45, ColabDock_average_DockQ: 0.56, example_4INS8_DockQ: GRASP 0.56 vs AFM 0.43 vs HADDOCK 0.18 vs ClusPro 0.06 vs ColabDock 0.33, CSP_average_DockQ_GRASP: 0.81 (4 cases), CSP_average_DockQ_AF M/HADDOCK/ClusPro/ColabDock: AFM 0.39 / HADDOCK 0.28 / ClusPro 0.21 / ColabDock 0.5, example_4G6M_DockQ_GRASP: 0.9, example_4G6J_DockQ_GRASP: 0.79, BM5.5_median_DockQ_GRASP: 0.64, BM5.5_success_rate_GRASP: 71.6%, Hitawala–Gray_median_DockQ_antibodies_GRASP: 0.477, Hitawala–Gray_median_DockQ_nanobodies_GRASP: 0.541, Hitawala–Gray_success_rate_antibodies_GRASP: 60.0%, Hitawala–Gray_success_rate_nanobodies_GRASP: 88.8%, AF3_second_best_median_DockQ_antibodies_AF3: 0.069, AF3_second_best_median_DockQ_nanobodies_AF3: 0.237, Experimental_DMS_median_DockQ_GRASP: 0.25, Experimental_DMS_success_rate_GRASP: 53.6%, Experimental_DMS_median_DockQ_AF3: 0.07, Experimental_DMS_success_rate_AF3: 39.3%, Combined_protocol_median_DockQ: 0.28, Combined_protocol_success_rate: 56.0%, mitochondria_predicted_PPIs_total: 144 PPIs predicted (121 had pLDDT > 75), XLs_satisfied_GRASP: 140/144 predicted PPIs satisfied XLs, XLs_satisfied_AFM: 31/144, median_TM_score_on_17_ground_truth_pairs_GRASP: 0.881, median_TM_score_on_17_ground_truth_pairs_AFM: 0.838, pLDDT_Pearson_correlation_with_DockQ: r = 0.39, pLDDT_Pearson_correlation_with_TM_score: r = 0.65, pLDDT_Pearson_correlation_with_LDDT: r = 0.87, improvements_in_pLDDT_vs_gains_in_DockQ_TM_LDDT: r = 0.38 (DockQ), 0.58 (TM), 0.77 (LDDT) |
| Application Domains | protein complex structure prediction / structural biology, antigen–antibody modelling and antibody design / immunotherapy, integrative structural biology (integrating XL-MS, CL, CSP, DMS, cryo-EM, NMR PRE, mutagenesis), in situ interactome modelling (mitochondrial PPI mapping), computational docking and restrained docking workflows |
374. De novo discovery of conserved gene clusters in microbial genomes with Spacedust, Nature Methods (September 15, 2025)
| Category | Items |
|---|---|
| Datasets | 1,308 representative bacterial genomes (reference database), Gold-standard BGC dataset (nine complete genomes), GTDB (Genome Taxonomy Database), AlphaFold structure database (and other structure DBs: PDB, ESMAtlas), PADLOC antiviral defense annotations, AntiSMASH functional annotation (version 8) |
| Models | T5, Transformer, Hidden Markov Model |
| Tasks | Clustering, Information Retrieval, Feature Extraction, Classification |
| Learning Methods | Representation Learning, Pre-training |
| Performance Highlights | AUC (precision–recall) i,i+1: 0.93, AUC (precision–recall) i,i+2: 0.93, AUC (precision–recall) i,i+3: 0.86, AUC (precision–recall) i,i+4: 0.81, AUC (precision–recall) i,i+1: 0.91, AUC (precision–recall) i,i+2: 0.89, AUC (precision–recall) i,i+3: 0.83, AUC (precision–recall) i,i+4: 0.77, PADLOC multi-gene defense clusters (reference): 5,520, Spacedust recovery of PADLOC clusters: 5,255 (95%), Spacedust full-length matches: 4,888 (93% of recovered clusters), Spacedust partial matches: 367 (7% of recovered clusters), Non-redundant clusters (paired matches grouped): 72,483 nonredundant clusters comprising 2.45M genes (58% of dataset), All pairwise cluster hits: 321.2M cluster hits in 106.6M cluster matches; mean genes per cluster match = 3.01, Spacedust assignment: 58% of all 4.2M genes assigned to conserved gene clusters; 35% of genes without any annotation assigned to clusters, Average F1 score (Spacedust): 0.61, Average F1 score (ClusterFinder): 0.44, Average F1 score (DeepBGC): 0.39, Average F1 score (GECCO): 0.43 |
| Application Domains | Microbial genomics (bacteria and archaea), Metagenomics / metagenome-assembled genomes, Microbiome research (environmental and human-associated microbiomes), Comparative genomics / evolutionary genomics (gene neighborhood conservation), Functional annotation of genes (operons, antiviral defense systems, biosynthetic gene clusters), CRISPR–Cas systems discovery (e.g., expansion of subtype III-E), Biosynthetic gene cluster discovery and natural product genome mining |
372. Guided multi-agent AI invents highly accurate, uncertainty-aware transcriptomic aging clocks, Preprint (September 12, 2025)
| Category | Items |
|---|---|
| Datasets | ARCHS4, ARCHS4 — blood subset, ARCHS4 — colon subset, ARCHS4 — lung subset, ARCHS4 — ileum subset, ARCHS4 — heart subset, ARCHS4 — adipose subset, ARCHS4 — retina subset |
| Models | XGBoost, LightGBM, Support Vector Machine, Linear Model, Transformer |
| Tasks | Regression, Feature Selection, Feature Extraction, Clustering |
| Learning Methods | Supervised Learning, Ensemble Learning, Imbalanced Learning |
| Performance Highlights | R2: 0.619, R2: 0.604, R2: 0.574, R2_Ridge: 0.539, R2_ElasticNet: 0.310, R2: 0.957, MAE_years: 3.7, R2_all: 0.726, MAE_all_years: 6.17, R2_confidence_weighted: 0.854, MAE_confidence_weighted_years: 4.26, mean_calibration_error: 0.7%, R2_per_window_range: ≈0.68–0.74, lung_R2: 0.969, blood_R2: 0.958, ileum_R2: 0.958, heart_R2: 0.910, adipose_R2: 0.887, retina_R2: 0.594 |
| Application Domains | aging biology / geroscience, transcriptomics, biomarker discovery, computational biology / bioinformatics, clinical biomarker development (biological age clocks), AI-assisted scientific discovery (multi-agent workflows) |
371. Flexynesis: A deep learning toolkit for bulk multi-omics data integration for precision oncology and beyond, Nature Communications (September 12, 2025)
| Category | Items |
|---|---|
| Datasets | CCLE, GDSC2, TCGA (multiple cohorts: pan-cancer, COAD, ESCA, PAAD, READ, STAD, UCEC, UCS, LGG, GBM), METABRIC, DepMap, ProtTrans protein sequence embeddings (precomputed), describePROT features, STRING interaction networks, METABRIC (as dataset entry repeated for clarity), Single-cell CITE-Seq of bone marrow, PRISM drug screening and CRISPR screens (DepMap components) |
| Models | Multi-Layer Perceptron, Feedforward Neural Network, Variational Autoencoder, Triplet Network, Graph Neural Network, Graph Convolutional Network, Support Vector Machine, Random Forest, XGBoost, Random Survival Forest, Graph Attention Network |
| Tasks | Regression, Classification, Survival, Clustering, Dimensionality Reduction, Feature Selection, Representation Learning |
| Learning Methods | Supervised Learning, Unsupervised Learning, Multi-Task Learning, Transfer Learning, Fine-Tuning, Hyperparameter Optimization, Contrastive Learning, Representation Learning, Domain Adaptation |
| Performance Highlights | Pearson_correlation_Lapatinib: r = 0.6 (p = 7.750175e-42), Pearson_correlation_Selumetinib: r = 0.61 (p = 3.873949e-50), AUC_MSI_prediction: AUC = 0.981, logrank_p: p = 9.94475168880626e-10, per-cell-line_correlation_distribution: median correlations shown per cell line (N=1064) with improvement when adding ProtTrans embeddings; exact medians not specified numerically in text, cross-domain_TCGA->CCLE_before_finetuning_F1: approx. 0.16, cross-domain_TCGA->CCLE_after_finetuning_F1: up to 0.8 |
| Application Domains | Precision oncology / cancer genomics, Pharmacogenomics (drug response prediction), Clinical genomics (survival prediction, biomarker discovery), Functional genomics (gene essentiality prediction, DepMap analyses), Multi-omics data integration (transcriptome, methylome, CNV, mutation), Proteomics / protein sequence analysis (ProtTrans embeddings, describePROT), Single-cell multi-omics (CITE-Seq cell type classification proof-of-concept), Bioinformatics tool development and benchmarking |
370. Biophysics-based protein language models for protein engineering, Nature Methods (September 11, 2025)
| Category | Items |
|---|---|
| Datasets | Rosetta simulated pretraining data (METL-Local), Rosetta simulated pretraining data (METL-Global), GFP (green fluorescent protein) experimental dataset, DLG4-Abundance (DLG4-A) experimental dataset, DLG4-Binding (DLG4-B) experimental dataset, GB1 experimental dataset, GRB2-Abundance (GRB2-A) experimental dataset, GRB2-Binding (GRB2-B) experimental dataset, Pab1 experimental dataset, PTEN-Abundance (PTEN-A) experimental dataset, PTEN-Activity (PTEN-E) experimental dataset, TEM-1 experimental dataset, Ube4b experimental dataset, METL Rosetta datasets (archived) |
| Models | Transformer, Linear Model, Convolutional Neural Network, Feedforward Neural Network, Attention Mechanism, Multi-Head Attention |
| Tasks | Regression, Synthetic Data Generation, Data Augmentation, Optimization, Representation Learning |
| Learning Methods | Pre-training, Fine-Tuning, Supervised Learning, Self-Supervised Learning, Transfer Learning, Feature Extraction, Zero-Shot Learning, Representation Learning |
| Performance Highlights | mean_Spearman_correlation: 0.91, in_distribution_mean_Spearman: 0.85, out_of_distribution_mean_Spearman: 0.16, mutation_extrapolation_avg_Spearman_range: ~0.70-0.78, ProteinNPT_Spearman: 0.65, METL-Local_Spearman: 0.59, supervised_models_avg_Spearman: >0.75, ProteinNPT_avg_Spearman: 0.67, typical_Spearman: <0.3, GB1_supervised_models_Spearman: >=0.55, GB1_METL-Local_METL-Global_Spearman: >0.7, METL-Bind_median_Spearman: 0.94, METL-Local_median_Spearman: 0.93, Linear_median_Spearman: 0.92, Regime_METL-Bind_Spearman: 0.76, Regime_METL-Local_Spearman: 0.74, Regime_Linear_Spearman: 0.56, designed_variants_with_measurable_fluorescence: 16/20, Observed_5-mutants_success_rate: 5/5 (100%), Observed_10-mutants_success_rate: 5/5 (100%), Unobserved_5-mutants_success_rate: 4/5 (80%), Unobserved_10-mutants_success_rate: 2/5 (40%), competitive_on_small_N: Linear-EVE and Linear sometimes competitive with METL-Local on small training sets (dataset dependent), general_performance: CNN baselines were generally outperformed by METL-Local; specific numbers in Supplementary Fig.7 |
| Application Domains | Protein engineering, Biophysics / molecular simulation, Computational protein design, Structural biology, Enzyme engineering (catalysis), Fluorescent protein engineering (GFP brightness), Stability and expression prediction |
369. Towards agentic science for advancing scientific discovery, Nature Machine Intelligence (September 10, 2025)
| Category | Items |
|---|---|
| Datasets | AFMBench (autonomous microscopy benchmark), METR (benchmark for long-horizon multi-step tasks), Crystallographic databases (generic structured resources), Gene ontologies (as structured resources), Chemical reaction networks (structured representations) |
| Models | Transformer, Attention Mechanism, Multi-Head Attention, Multi-Layer Perceptron |
| Tasks | Experimental Design, Novelty Detection, Text Generation, Language Modeling, Sequence-to-Sequence, Graph Generation, Question Answering, Novelty Detection |
| Learning Methods | Active Learning, Transfer Learning, Self-Supervised Learning, Reinforcement Learning, Fine-Tuning, Supervised Learning |
| Performance Highlights | qualitative_outcome: struggles / compounds errors over time, qualitative_outcome: revealed critical failure modes |
| Application Domains | Chemistry, Materials Science, Microscopy / Laboratory Automation, Crystallography / Materials Characterization, Biology / Genomics (via gene ontologies), Clinical domains (clinical diagnosis mentioned as boundary condition), Social Sciences (noted as challenging domain), Autonomous Laboratories / Robotics-enabled synthesis |
368. Molecular-dynamics-simulation-guided directed evolution of flavoenzymes for atroposelective desaturation, Nature Synthesis (September 10, 2025)
| Category | Items |
|---|---|
| Datasets | MD simulation trajectories (monomer and tetramer), Docking inputs (substrate enantiomers into AlphaFold2-generated structure), X-ray crystal structures and small-molecule crystal data, Experimental enzymatic dataset: kinetics, yields and enantioselectivities (substrate scope), In-house enzyme collection (purified flavoenzymes panel) |
| Models | Hidden Markov Model |
| Tasks | Clustering, Experimental Design |
| Learning Methods | Unsupervised Learning |
| Performance Highlights | transition_time_ADes-1_vs_ADes-5: ADes-1 transition time from macrostate A to B was an order of magnitude longer than in ADes-5, transition_probability_ADes-1_vs_ADes-5: transition probability from the initial state to the final state in the ADes-1 system was an order of magnitude smaller than in ADes-5, kcat_ADes-1: (1.6 ± 0.1) × 10^-3 min^-1, KM_ADes-1: 1.7 ± 0.3 mM, kcat_ADes-5: (9.3 ± 1.6) × 10^-2 min^-1, KM_ADes-5: 1.3 ± 0.4 mM, kcat/KM_improvement: 70-fold improvement in kcat/KM for ADes-5 relative to ADes-1, yield_best: >99% (selected substrates), ee_best: >99% e.e. (selected substrates), yield_ADes-5_for_2a: 98% yield with 89% e.e. (ADes-5 produced 98% yield of desired product with 89% e.e. in one round; later preparative scale 93% yield, 87% e.e.) |
| Application Domains | Biocatalysis, Enzyme engineering / directed evolution, Computational structural biology / molecular dynamics, Synthetic organic chemistry (atroposelective synthesis of biaryls), Drug discovery (synthesis of pharmaceutically relevant atropisomers) |
367. RoboChemist: Long-Horizon and Safety-Compliant Robotic Chemical Experimentation, Preprint (September 10, 2025)
| Category | Items |
|---|---|
| Datasets | RoboChemist collected VLA fine-tuning dataset, Training configurations for inner-loop enhancement experiments, Referenced external benchmarks and datasets (not directly used for training) |
| Models | Transformer, Diffusion Model, GPT |
| Tasks | Control, Planning, Policy Learning, Object Localization, Decision Making |
| Learning Methods | Fine-Tuning, Prompt Learning, Supervised Learning, Transfer Learning, Batch Learning |
| Performance Highlights | Overall average success rate improvement vs VLA baselines (%): 23.57, Overall average compliance rate increase: 0.298, Grasp Glass Rod SR (%): 55, Grasp Glass Rod CR: 0.325, Grasp Glass Rod SR (%): 20, Grasp Glass Rod CR: 0.100, Grasp Glass Rod SR (%): 40, Grasp Glass Rod CR: 0.200, Grasp Glass Rod SR (%): 85, Grasp Glass Rod CR: 0.750, Grasp Glass Rod SR (%): 95, Grasp Glass Rod CR: 0.875, Heat Platinum Wire SR (%): 20, Heat Platinum Wire CR: 0.063, Heat Platinum Wire SR (%): 60, Heat Platinum Wire CR: 0.363, Heat Platinum Wire SR (%): 55, Heat Platinum Wire CR: 0.325, Heat Platinum Wire SR (%): 70, Heat Platinum Wire CR: 0.575, Heat Platinum Wire SR (%): 90, Heat Platinum Wire CR: 0.800, Insert into Solution SR (%): 10, Insert into Solution CR: 0.050, Insert into Solution SR (%): 80, Insert into Solution CR: 0.775, Insert into Solution SR (%): 80, Insert into Solution CR: 0.800, Insert into Solution SR (%): 85, Insert into Solution CR: 0.850, Insert into Solution SR (%): 95, Insert into Solution CR: 0.950, Pour Liquid SR (%): 25, Pour Liquid CR: 0.288, Pour Liquid SR (%): 90, Pour Liquid CR: 0.675, Pour Liquid SR (%): 80, Pour Liquid CR: 0.475, Pour Liquid SR (%): 80, Pour Liquid CR: 0.663, Pour Liquid SR (%): 95, Pour Liquid CR: 0.800, Stir the Solution SR (%): 15, Stir the Solution CR: 0.075, Stir the Solution SR (%): 75, Stir the Solution CR: 0.400, Stir the Solution SR (%): 85, Stir the Solution CR: 0.600, Stir the Solution SR (%): 95, Stir the Solution CR: 0.650, Stir the Solution SR (%): 100, Stir the Solution CR: 0.825, Transfer the Solid SR (%): 15, Transfer the Solid CR: 0.063, Transfer the Solid SR (%): 75, Transfer the Solid CR: 0.513, Transfer the Solid SR (%): 80, Transfer the Solid CR: 0.525, Transfer the Solid SR (%): 85, Transfer the Solid CR: 0.538, Transfer the Solid SR (%): 95, Transfer the Solid CR: 0.675, Press the Button SR (%): 0, Press the Button CR: 0.100, Press the Button SR (%): 65, Press the Button CR: 0.413, Press the Button SR (%): 70, Press the Button CR: 0.575, Press the Button SR (%): 75, Press the Button CR: 0.613, Press the Button SR (%): 85, Press the Button CR: 0.663, Complete task: Mix NaCl and CuSO4 SR (%): 80, Complete task: Mix NaCl and CuSO4 CR: 0.450, Complete task: Mix NaCl and CuSO4 SR (%): 95, Complete task: Mix NaCl and CuSO4 CR: 0.775, Visual prompting comparison (Grasp Glass Rod) SR (π0) (%): 40, Visual prompting comparison (Grasp Glass Rod) CR (π0): 0.200, ReKep+π0 SR: 35, ReKep+π0 CR: 0.200, MOKA+π0 SR: 65, MOKA+π0 CR: 0.350, RoboChemist w/o CL SR: 85, RoboChemist w/o CL CR: 0.750, Config 1 average SR (%): 67.14, Config 2 average SR (%): 70.00, Config 3 average SR (%): 62.14, Config 4 average SR (%): 22.14, Pouring two cups (standalone VLA w/o visual prompt) success count: 16/20, Pouring three cups (standalone VLA w/o visual prompt) success count: 5/20, Pouring two cups (with visual prompt) success count: 19/20, Pouring three cups (with visual prompt) success count: 17/20 |
| Application Domains | Robotic chemistry / laboratory automation, Robotic manipulation (bimanual) in hazardous and deformable-material settings, Vision-language-grounded robotic control, Long-horizon safe procedural automation in chemistry experiments |
366. AI mirrors experimental science to uncover a mechanism of gene transfer crucial to bacterial evolution, Cell (September 09, 2025)
| Category | Items |
|---|---|
| Datasets | None |
| Models | None |
| Tasks | None |
| Learning Methods | None |
| Performance Highlights | None |
| Application Domains | unspecified / unclear (paper text is heavily corrupted/encoded and does not explicitly state an application domain), possible domain-specific experimental science (terms such as FO!m{/�mtoz�t��, yomskzt�y, WOIO, SST appear repeatedly but do not match standard AI dataset/model/task names) |
365. SurFF: a foundation model for surface exposure and morphology across intermetallic crystals, Nature Computational Science (September 09, 2025)
| Category | Items |
|---|---|
| Datasets | Design space of generated intermetallic surfaces, SurFF training dataset (active learning-generated), Active learning test set, In-distribution (ID) test set, Out-of-distribution (OOD) test set, Large-scale prediction dataset (predictions performed on catalytic materials), Experimental validation dataset (literature + original experiments) |
| Models | Graph Neural Network, Graph Convolutional Network, Transformer, Attention Mechanism, Ensemble Learning |
| Tasks | Regression, Optimization, Multi-class Classification, Representation Learning, Clustering / Sampling (Active Learning selection - diversity sampling) |
| Learning Methods | Supervised Learning, Active Learning, Ensemble Learning, Pre-training, Fine-Tuning, Transfer Learning, Representation Learning |
| Performance Highlights | MAE_active_learning_test_set (surface energy, meV Å−2): 3.8, MAE_ID_test_set (surface energy, meV Å−2): 3.0, MAE_OOD_test_set (surface energy, meV Å−2): 10.5, Structural_RMS_error_between_SurFF_and_DFT_relaxed_structures (Å): 0.109, Speedup_vs_DFT: 105x, Active_learning_dataset_size_generated (surface energy datapoints): 12,000 (final reported training set), DFT_compute_cost_for_dataset (CPU-hours): 155,612, Overall_synthesizability_accuracy (SurFF vs DFT / experimental context): 71.9%, High_synthesizability_accuracy (SurFF, ID): 77.1%, OOD_top-5_accuracy (surface synthesizability): 0.810, OOD_high_accuracy: 0.744, Experimental_facets_predicted_accuracy: 73.1%, Improvement_single_point_energy_after_fine_tuning (%): 45, Improvement_single_point_force_after_fine_tuning (%): 53, Improvement_surface_energy_MAE_after_fine_tuning (%): 35, Improvement_top-3_accuracy_after_fine_tuning (%): 5.1 |
| Application Domains | Heterogeneous catalysis / catalyst design, Materials discovery and screening (intermetallic crystals), Computational materials science / surface science, Quantum chemistry acceleration (DFT surrogate), Nanoparticle morphology prediction (Wulff construction) |
363. AI-driven protein design, Nature Reviews Bioengineering (September 08, 2025)
| Category | Items |
|---|---|
| Datasets | UniProt, Protein Data Bank (PDB), AlphaFoldDB / AlphaFold-predicted structures, ESM Metagenomic Atlas (ESM-predicted structures), ESM-IF training set (AlphaFold2-predicted structures), CodonTransformer training set, AAV in silico library / experimental validation sets (AAV capsid case study), De novo luciferase design library (NTF2-like scaffolds), Variational Synthesis generative synthesis scale |
| Models | Transformer, Recurrent Neural Network, Convolutional Neural Network, Graph Neural Network, Message Passing Neural Network, Diffusion Model, Variational Autoencoder, Denoising Diffusion Probabilistic Model, Attention Mechanism / Self-Attention Network, Graph Convolutional Network, Geometric 3D networks |
| Tasks | Language Modeling, Clustering, Ranking, Binary Classification, Regression, Data Generation, Synthetic Data Generation |
| Learning Methods | Supervised Learning, Unsupervised Learning, Reinforcement Learning, Transfer Learning, Zero-Shot Learning, Few-Shot Learning, Active Learning, Pre-training, Fine-Tuning, Contrastive Learning |
| Performance Highlights | sequence_recovery: from 41.2% to >50.0%, experimental_success: high success rates across diverse, experimentally validated design settings (qualitative), antibody_evolution_binding_improvement: up to 160-fold (some immature antibodies); up to 7-fold (four mature antibodies), filtered_candidates: 201,426 from ~1e10 virtual library, experimental_viable_count: 110,689, viability_rate: 58.1%, activity_improvement: up to 100-fold improvement in protein activity across diverse targets (EVOLVEpro study referenced), binding_affinity_increase: up to 37-fold (rational antibody optimization using ESM-IF), binder_design_affinity: nanomolar affinities reported in de novo binder designs (Gainza et al.; qualitative), MaSIF speedup: searches 20–200 times faster than conventional docking (MaSIF description) |
| Application Domains | Protein engineering / protein design, Drug discovery (therapeutic protein and binder design), Biotechnology (industrial enzymes, developability improvement), Synthetic biology (design of novel proteins and biological systems), Gene therapy (AAV capsid engineering), Antibody therapeutics (optimization, de-immunization, humanization), Structural biology (structure prediction and complex modelling), DNA synthesis and experimental protocol optimization |
362. Accelerating protein engineering with fitness landscape modelling and reinforcement learning, Nature Machine Intelligence (September 08, 2025)
| Category | Items |
|---|---|
| Datasets | UniRef50 (UR50), ProteinGym v0.1 (substitution), ProteinGym Indel, FLIP (including FLIP AAV VP1 splits), FLEXS benchmark (five ground-truth landscapes), TEM-1 single-mutant DMS (Stiffler et al. ref. 45), Curated ESBL dataset (clinical variants), TEM-1 wet-laboratory validation set (this study), One-to-multi evaluation datasets (GB1, TEM-1, Pab1, PSD-95, GRB2, APP, GFP, YAP1, VP1) |
| Models | μFormer, μSearch, Transformer, Multi-Layer Perceptron, Convolutional Neural Network, Multi-Head Attention / Self-Attention Network / Attention Mechanism, Ridge (one-hot-embedding-based), ECNet / DeepSequence / EVmutation / ESM / Tranception / ProteinNPT / ConFit / Augmented DeepSequence |
| Tasks | Regression, Ranking, Binary Classification, Optimization, Policy Learning, Out-of-Distribution Learning, Decision Making |
| Learning Methods | Self-Supervised Learning, Pre-training, Fine-Tuning, Supervised Learning, Transfer Learning, Reinforcement Learning, Policy Gradient, Actor-Critic, Representation Learning |
| Performance Highlights | Spearman_rho_>0.7percentage: over 50% of ProteinGym datasets, Spearman_rho>0.9_datasets: 6 datasets exceeded 0.9, indel_Spearman_rho: above 0.8 for Mut-Des and Des-Mut settings on FLIP AAV / ProteinGym Indel, top-100_recall_average: 0.165 (single-to-multi setting across nine proteins), top-500_recall_average: 0.341 (single-to-multi setting across nine proteins), auPRC_examples: auPRC used for ultrahigh-value mutants (GB1 n=681, GRB2 n=426, YAP1 n=488, VP1 n=131) — μFormer outperforms alternatives, Spearman_MIC_correlation_TEM-1: reported values: ρ = 0.94 (text) and ρ = 0.92 (figure caption) with MIC measurements, oracle_queries_for_superior_scores: μSearch reached fitness levels unreachable by other algorithms by ~50,000 queries (single-round TEM-1 with μFormer oracle); comparative experiments with 250,000 queries per method, high_score_sequences_identified: for predicted score >0.3, μSearch identified over 2,000 sequences that others failed to detect, sample_efficiency: μSearch demonstrated superior sample efficiency on several protein landscapes in FLEXS benchmarks (multi-round setting), computational_budget_setting: multi-round simulated design: 10 rounds × 100 candidates per round, budget 5,000 local approximate model calls, computational_screened: 1,000,000 mutant sequences screened across six μSearch runs, candidates_selected_for_experiment: top-200 variants selected for E. coli growth assay, experimental_success_rate: 47 distinct RL-designed variants (23.5% of top-200 tested) exhibited superior growth on cefotaxime relative to wild type; random baseline: 12% improved, growth_fold_change: E. coli harbouring certain variants exhibited growth rates up to 2,000-fold higher than wild type, novel_high_activity_variant: G236S;T261V surpassed activity level of known quadruple mutant A40G;E102K;M180T;G236S |
| Application Domains | Protein engineering, Enzyme optimization (e.g., TEM-1 β-lactamase activity against cefotaxime), Antibody design, Drug-resistance prediction, Machine-guided directed evolution / sequence design |
361. AI-Driven Defect Engineering for Advanced Thermoelectric Materials, Advanced Materials (September 04, 2025)
| Category | Items |
|---|---|
| Datasets | Gaultois et al. dataset, Na et al. dataset, Sierepeklis / ChemDataExtractor auto-generated TE dataset, SpringerMaterials TE dataset, Itani et al. (LLM-extracted) TE dataset, Chen et al. high-throughput DFT TE dataset, Petretto et al. phonon database, Ricci et al. electronic transport dataset, Toher et al. phonon thermal conductivity dataset, Open Quantum Materials Database (OQMD), Materials Project (MP) |
| Models | Gradient Boosting Tree, XGBoost, LightGBM, Gaussian Process, Support Vector Machine, Random Forest, Multi-Layer Perceptron, Convolutional Neural Network, ResNet, Transformer, Attention Mechanism, Variational Autoencoder, Generative Adversarial Network, Diffusion Model, BERT, GPT, Graph Neural Network, Message Passing Neural Network, Graph Convolutional Network, Gaussian Process, Variational Autoencoder, Feedforward Neural Network |
| Tasks | Regression, Feature Selection, Clustering, Representation Learning, Data Generation, Synthetic Data Generation, Active Learning, Clustering, Hyperparameter Optimization / Optimization, Clustering |
| Learning Methods | Supervised Learning, Unsupervised Learning, Self-Supervised Learning, Active Learning, Transfer Learning, Generative Learning, Representation Learning, Feature Selection, Fine-Tuning |
| Performance Highlights | MAE_zT: 0.06, MAE_Seebeck_μV_per_K: 49, zT_enhancement_percent: up to 104%, thermal_conductivity_change_percent: ≈14% (reported in specific Pb–Se–Te–S local-order study) |
| Application Domains | Thermoelectrics, Energy materials, Materials science and engineering, Computational materials discovery / high-throughput screening, Atomistic simulation (DFT, MD) accelerated by ML, Defect engineering (point defects, dislocations, grain boundaries, interfaces), High-entropy alloys / high-entropy thermoelectrics, Sustainability-aware materials design |
359. Supervised learning in DNA neural networks, Nature (September 03, 2025)
| Category | Items |
|---|---|
| Datasets | Modified National Institute of Standards and Technology (MNIST) — subsets converted to 100-bit binary patterns |
| Models | Feedforward Neural Network |
| Tasks | Binary Classification, Image Classification, Clustering |
| Learning Methods | Supervised Learning, Batch Learning |
| Performance Highlights | activated_memory_classification_correctness: Fluorescence kinetics experiments confirmed correct outputs for all selected test patterns on activated memories (12 tests per class) (Fig. 3), representative_tests_after_training: 72 representative tests classified successfully after 3 distinct training processes (Fig. 6d; simulations and experiments shown), lower_bound_accuracy_estimates_on_full_MNIST_subsets: examples: 56% for threes, 46% for fours, 56% for sixes, 71% for sevens (expected lower bounds for full dataset given 20% diagonal margin); Extended Data Fig.1: 53% for ‘0’ and 83% for ‘1’ as lower bounds, classification_time_scale: Typical testing fluorescence kinetics monitored up to 8 hours; reporting often shown at 4–8 h endpoints, system_size: 100-bit, 2-memory network involved >700 distinct species in a single test tube and >1,200 unique strands across learning and testing, learned_weight_readout_noise: Before training, background signals in memories 1 and 2 were 0.4 ± 0.4% and 0.7 ± 0.3% of total signal; after learning, pixels with 0 values in all 10 training patterns showed low signals (0.5 ± 0.3% for ‘1’ memory, 1.1 ± 0.4% for ‘0’), learned_signal_enrichment: Other pixels showed up to 13-fold signal increases over background in learned weights (endpoint values), successful_memory_integration: Learned weights consistently stored across different training orders; memory integration robust |
| Application Domains | Molecular computation (DNA strand-displacement circuits), Synthetic biology / DNA nanotechnology, Bioengineering (cell-free molecular devices), Diagnostics (molecular classifiers for disease biomarkers, potential future applications), Programmable soft materials and active materials (materials that adapt using DNA circuits) |
358. Machine learning in X-ray diffraction for materials discovery and characterization, Matter (September 03, 2025)
| Category | Items |
|---|---|
| Datasets | None |
| Models | None |
| Tasks | None |
| Learning Methods | None |
| Performance Highlights | None |
| Application Domains | None |
357. A generalizable pathology foundation model using a unified knowledge distillation pretraining framework, Nature Biomedical Engineering (September 02, 2025)
| Category | Items |
|---|---|
| Datasets | GPFM pretraining collection (56 sources), TCGA (multiple cohorts, e.g., TCGA-BRCA, TCGA-LUAD, TCGA-GBMLGG, etc.), CPTAC (e.g., CPTAC-LUAD), PANDA, CAMELYON16 + CAMELYON17 (CAMELYON), CRC-100K (NCT-CRC-HE-100K + CRC-VAL-HE-7K), CRC-MSI, CCRCC-TCGA-HEL, PanCancer-TCGA (PanCancer-TCGA images), PanCancer-TIL, PCAM, WSSS4LUAD, BACH, BreakHis, UniToPatho, PathVQA, WSI-VQA, TCGA WSI-Report, PatchGastricADC22, UBC-OCEAN, GasHisDB, CHA / ESCA dataset (esophageal carcinoma subtyping), IMP-CRS (colon lesion grading), HANCOCK, EBRAINS (Digital Tumour Atlas), Center-1 to Center-5 cohorts |
| Models | Vision Transformer, ResNet, BERT, Transformer, Attention Mechanism, Linear Model, Encoder-Decoder |
| Tasks | Image Classification, Survival Analysis, Image Retrieval, Question Answering, Text Generation, Binary Classification, Multi-class Classification |
| Learning Methods | Self-Supervised Learning, Knowledge Distillation, Pre-training, Fine-Tuning, Multi-Instance Learning, Contrastive Learning, Transfer Learning, Representation Learning |
| Performance Highlights | average_rank_WSIs_across_36_tasks: 1.22, AUC_overall: 0.891, balanced_accuracy: 0.752, weighted_F1: 0.736, AUC_internal: 0.938, AUC_external: 0.832, average_rank_survival_15_tasks: 2.1, average_C-index: 0.665, average_rank_ROI_16_tasks: 1.88, average_AUC: 0.946, weighted_F1: 0.865, balanced_accuracy: 0.866, top-1_accuracy: 0.906, top-3_accuracy: 0.993, top-5_accuracy: 0.995, patch_level_rank: second-best (close to CONCH), WSI_level_metrics: best or second-best across 6 out of 7 metrics (detailed per-metric results in Supplementary Table 39), ranking: Phikon best; GPFM second-best across BLEU/METEOR/ROUGE-L metrics on TCGA WSI-Report and PatchGastricADC22, TP53_AUC_LUAD: 0.855, IDH1_AUC_internal: 0.986, IDH1_AUC_external: 0.943, average_performance_across_72_tasks: 0.749 |
| Application Domains | Computational Pathology, Digital Histopathology, Cancer Diagnosis and Subtyping, Mutation Prediction from Histology, Survival Prognosis Prediction, Medical Image Retrieval, Pathology Visual Question Answering (clinical decision support), Automated Pathology Report Generation |
356. PXDesign: Fast, Modular, and Accurate De Novo Design of Protein Binders, Preprint (September 02, 2025)
| Category | Items |
|---|---|
| Datasets | Cao data, RFDiffusion wet-lab set, EGFR challenge / EGFR competition dataset, SKEMPI subset (filtered), PDB subset (curated up to May 1, 2021) + AFDB + MGnify distillation, AlphaProteo (published baselines / datasets referenced), RFpeptides / cyclic peptide targets (benchmarks) |
| Models | Diffusion Model, Denoising Diffusion Probabilistic Model, Transformer, Multi-Layer Perceptron |
| Tasks | Synthetic Data Generation, Binary Classification, Regression, Ranking, Clustering, Data Generation |
| Learning Methods | Generative Learning, End-to-End Learning, Backpropagation, Gradient Descent, Ensemble Learning, Pre-training, Fine-Tuning, Hyperparameter Optimization, End-to-End Learning |
| Performance Highlights | nanomolar_hit_rates_by_target_Table1: {‘PD-L1’: ‘72.7%’, ‘VEGF-A’: ‘47.1%’, ‘SC2RBD’: ‘50.0%’, ‘TrkA’: ‘20.0%’, ‘TNF-α’: ‘0.0%’}, summary: nanomolar binder hit rates of 20–73% across five of six targets (PXDesign overall), in_silico_comparative: PXDesign-h shows strong in silico performance and is competitive with hallucination baselines; slower but superior on specific cyclic peptide targets (e.g., MDM2, MCL1, IL17A, TNF-α)., 24h_yield_comparison: PXDesign-d delivers more successful designs within 24h than any hallucination method (PXDesign-h, BindCraft, BoltzDesign1) due to faster generation and higher pass rates (no exact numeric given)., RFDiffusion_re-ranking: Re-ranking the 95 RFdiffusion designs by Protenix ipTM (top10/top15) substantially increases observed success rates (Figure 1d)., AUC_AP_filtering: Protenix-derived scores outperform AF2-IG across most targets in AUC and Average Precision on Cao data (Figure 8); Protenix matches/exceeds AF3 on SKEMPI subset (Figure 1f)., sequence_design_protocol: ProteinMPNN-CA used for sequence design with default settings; for diffusion designs one sequence per backbone; for hallucination designs 8 sequences per structure., impact_on_success: Design considered successful if at least one sequence meets filter criteria; protocol differences accounted for in comparisons (no single numeric). |
| Application Domains | protein–protein binder design (de novo binder design), unconditional protein monomer design (de novo protein backbone generation), cyclic peptide binder design, nucleic acid (RNA/DNA) binder design (case studies; qualitative), small-molecule binder design (case studies; qualitative), post-translationally modified protein design (case studies; qualitative) |
355. Developing machine learning for heterogeneous catalysis with experimental and computational data, Nature Reviews Chemistry (September 2025)
| Category | Items |
|---|---|
| Datasets | Open Catalyst (OC20 / OC22 / OCx24), CatalysisHub / IoChem-BD / NOMAD / AFLOW / OQMD / Materials Project / BEAST DB, Gregoire scanning droplet / combinatorial experimental screening datasets, Taniike high-throughput OCM screening, Yildirim methane reforming compilations, Zavyalova OCM literature database, Perovskite / pseudo-quaternary combinatorial OER datasets, Large computational screening datasets (examples reported), IrO2 / IrO3 polymorph DFT dataset (Flores et al.) |
| Models | Linear Model, Decision Tree, Random Forest, Gradient Boosting Tree, XGBoost, LightGBM, Support Vector Machine, Gaussian Process, Multi-Layer Perceptron, Convolutional Neural Network, Graph Neural Network, K-means, Principal Component Analysis, Random Forest |
| Tasks | Regression, Classification, Clustering, Dimensionality Reduction, Feature Extraction, Clustering, Optimization, Experimental Design, Clustering, Representation Learning |
| Learning Methods | Supervised Learning, Unsupervised Learning, Active Learning, Transfer Learning, Semi-Supervised Learning, Reinforcement Learning, Unsupervised Learning |
| Performance Highlights | acceleration_vs_random: up to 20-fold |
| Application Domains | Heterogeneous catalysis, Electrocatalysis (water splitting, OER, HER, ORR), CO and CO2 conversion and reduction, Methane reforming and methane reactivity (oxidative coupling of methane, methanation), Nitrogen reactivity (N2 reduction, NO decomposition, ammonia synthesis), High-throughput experimental catalyst screening (combinatorial materials), Computational high-throughput DFT screening / materials discovery |
354. Protein evolution as a complex system, Nature Chemical Biology (September 2025)
| Category | Items |
|---|---|
| Datasets | vast quantities of protein sequence data (unspecified), synthetic datasets generated by ancestral sequence reconstruction, in silico-evolved variant populations |
| Models | Graph Neural Network, Autoencoder, Variational Autoencoder, Transformer, BERT |
| Tasks | Language Modeling, Regression, Sequence-to-Sequence, Dimensionality Reduction, Optimization, Decision Making, Data Generation, Clustering, Representation Learning |
| Learning Methods | Self-Supervised Learning, Reinforcement Learning, Representation Learning, Manifold Learning, Pre-training, Generative Learning |
| Performance Highlights | None |
| Application Domains | protein evolution, protein engineering and design, molecular evolution, directed evolution, synthetic biology, complex systems modeling, in silico evolutionary simulation, fitness prediction / protein fitness landscapes |
353. Robot-assisted mapping of chemical reaction hyperspaces and networks, Nature (September 2025)
| Category | Items |
|---|---|
| Datasets | Full robot-generated spectral dataset (all automated reactions), E1 hyperspace cube, SN1 hyperspace cube (9-butyl-9H-fluoren-9-ol), SN1 hyperspace cube (15a substrate with anomaly example), Ugi-type four-component hyperspace, Hantzsch reaction hyperspace, Prussian blue analogue (PBA) catalyst composition space |
| Models | Linear Model, Radial Basis Function Network |
| Tasks | Feature Extraction, Anomaly Detection, Regression, Optimization, Clustering |
| Learning Methods | Maximum Likelihood Estimation |
| Performance Highlights | yield_estimate_uncertainty_example: ±1% absolute (20% yield reported spread to 19–21%), optical_vs_purified_R2: 0.96, optical_repeatability_RSD: 2% (n=54, same crude mixture), workflow_repeatability_RSD: 5% (n=27, entire workflow), SN1_thermodynamic_parameters_estimates: ΔH = -30.7 ± 1.4 kJ mol−1 (SN1); other parameters for E1: values reported in Methods, fit_quality_examples: Kinetic models fitted closely to experimental hyperspace data (Fig. 2e,f; Fig. 4e–h) |
| Application Domains | organic chemistry / synthetic reaction discovery, reaction optimization, analytical spectroscopy (UV-Vis spectral analysis), catalysis (Prussian blue analogues for styrene epoxidation), materials chemistry (PBA composition screening), chemical reaction network reconstruction and mechanistic studies |
352. Electron flow matching for generative reaction mechanism prediction, Nature (September 2025)
| Category | Items |
|---|---|
| Datasets | USPTO-full (processed), FlowER mechanistic dataset (authors’ curated), Pistachio (subset of patent reactions not assigned a reaction class), RMechDB (supplemental), PMechDB (supplemental) |
| Models | Transformer, Graph Neural Network, Radial Basis Function Network, Encoder-Decoder |
| Tasks | Sequence-to-Sequence, Structured Prediction, Graph Generation |
| Learning Methods | Generative Learning, Denoising Diffusion Probabilistic Model, Supervised Learning, Pre-training, Fine-Tuning, Representation Learning |
| Performance Highlights | validity_rate: ≈95% (valid intermediate or product SMILES strings generated in approximately 95% of test reactions), model_size_default_params: 7 million parameters (default FlowER), model_size_large_params: 16 million parameters (FlowER-large), SMILES_validity: 70.2%, heavy_atom_conservation: 39.1%, cumulative_conservation_(heavy_atom+proton+electron): 33.0%, model_size: 12 million parameters, SMILES_validity_G2S: 76.3%, SMILES_validity_G2S+H: 78.8%, heavy_atom_conservation_G2S: 30.7%, heavy_atom_conservation_G2S+H: 27.7%, cumulative_conservation_G2S: 17.2%, cumulative_conservation_G2S+H: 19.0%, model_size: 18 million parameters (G2S), few_shot_fine_tune_500steps_top1_step_accuracy_FlowER: ≈35%, few_shot_fine_tune_500_steps_top10_step_accuracy_FlowER: ≈40%, G2S_performance_on_500_steps: near-zero (reported as near-zero performance by G2S), fine_tune_32_examples_top1_pathway_accuracy>=65%_count: 9 out of 12 unseen reaction types, recovered_products_from_Pistachio_22k: 351 products recovered from 22,000 unrecognized reactions |
| Application Domains | medicinal chemistry / synthetic planning, materials discovery, combustion chemistry, atmospheric chemistry, electrochemical systems, automated quantum chemical calculations / thermodynamic and kinetic feasibility estimation, reaction design and predictive chemistry |
351. The Biodiversity Cell Atlas: mapping the tree of life at cellular resolution, Nature (September 2025)
| Category | Items |
|---|---|
| Datasets | Earth BioGenome Project genomes, Biodiversity Cell Atlas (BCA) single-cell datasets (planned/curated by this paper), Human Cell Atlas, Tabula Muris (single-cell transcriptomics of 20 mouse organs), Fly Cell Atlas, Malaria Cell Atlas, Stony coral cell atlas, GoaT (Genomes on a Tree) metadata, TranscriptFormer preprint / cross-species generative cell atlas (preprint dataset/model) |
| Models | Convolutional Neural Network, Transformer, Variational Autoencoder, Graph Neural Network |
| Tasks | Regression, Language Modeling, Clustering, Dimensionality Reduction, Feature Extraction, Clustering, Embedding Learning, Generative Learning |
| Learning Methods | Supervised Learning, Pre-training, Transfer Learning, Fine-Tuning, Self-Supervised Learning, Generative Learning, Representation Learning, Embedding Learning |
| Performance Highlights | None |
| Application Domains | Comparative genomics and evolutionary biology, Single-cell biology and cell atlas construction, Biotechnology, Biomedicine (disease variant prediction, functional genomics), Environmental science and biomonitoring, Synthetic biology (design of regulatory sequences and circuits), Computational method and software infrastructure (data standards, pipelines) |
350. Generalizable descriptors for automatic titanium alloys design by learning from texts via large language model, Acta Materialia (September 01, 2025)
| Category | Items |
|---|---|
| Datasets | Data S1 (corpus of abstracts), Data S2, Data S3, Data S4, Unexplored candidate space (generated search set), GitHub repository data and code |
| Models | Transformer, BERT, Linear Model, Random Forest, Gradient Boosting Tree, Support Vector Machine, Gaussian Process, Multi-Layer Perceptron, Genetic Algorithm |
| Tasks | Language Modeling, Representation Learning, Regression, Dimensionality Reduction, Clustering, Binary Classification |
| Learning Methods | Self-Supervised Learning, Pre-training, Supervised Learning, Transfer Learning, Contrastive Learning, Evolutionary Learning, Mini-Batch Learning, Representation Learning, Batch Learning |
| Performance Highlights | pretraining_loss: 0.48, vocabulary_size: 4462 tokens, training_samples: >780,000, improvement_over_conventional_descriptors_tensile_strength_R2: 8.04% (GBR improvement reported), improvement_over_conventional_descriptors_yield_strength_R2: 5.91% (GBR improvement reported), improvement_over_conventional_descriptors_elongation_R2: 8.24% (GBR improvement reported), improvement_over_conventional_descriptors_tensile_strength_R2: 44.2%, improvement_over_conventional_descriptors_yield_strength_R2: 25.3%, improvement_over_conventional_descriptors_elongation_R2: 29.8%, improvement_over_conventional_descriptors_yield_strength_R2: 19.3%, improvement_over_conventional_descriptors_elastic_modulus_R2: 7.25%, evaluation_metrics_used: R2, MAE, RMSE (five-fold cross-validation), evaluation_metrics_used: R2, MAE, RMSE (five-fold cross-validation), evaluation_metrics_used: R2, MAE, RMSE (five-fold cross-validation), relative_performance: Paper reports that their model’s embeddings generally outperform SciBERT, MatBERT, MatSciBERT across tested cases (see Table S5 for details), GA_parameters: population=800, generations=450, crossover_rate=80%, mutation_rate=5%, result: Dimensionality reduced to 13 optimized descriptors; fitness (R2) converged by generation 450 for GBR |
| Application Domains | Materials science (titanium alloys design), Metallurgy, Aerospace materials (undercarriage, structural applications), Biomedical implants (low-modulus titanium alloys), Materials informatics / text-mining for materials design |
349. From Automation to Autonomy: A Survey on Large Language Models in Scientific Discovery, Preprint (August 30, 2025)
| Category | Items |
|---|---|
| Datasets | LitQA / PaperQA, ArxivDIGESTables, arXiv2Table, Text-Tuple-Table, TKGT, IdeaBench / AI Idea Bench 2025 / LiveIdeaBench, MOOSE-Chem, BioPlanner benchmark, ARCADE, DS-1000, MLE-Bench / MLE-Bench (MLE-Bench), AIDE, Chain-of-Table, TableBench, ChartQA, CharXiv / ChartX & ChartVLM, AutomaTikZ, Text2Chart31, ClaimCheck, SciReplicate-Bench, LLM-SRBench / LLM-SR, Gravity-Bench-v1, InfiAgent-DABench, BLADE / DiscoveryBench / DiscoveryWorld / ScienceAgentBench / CURIE / EAIRA / ResearchBench |
| Models | Transformer, Vision Transformer, Graph Neural Network, Multi-Layer Perceptron |
| Tasks | Information Retrieval, Text Summarization, Text Generation, Planning, Question Answering, Regression, AutoML, Image Generation, Survival Analysis, Language Modeling, Hyperparameter Optimization |
| Learning Methods | Reinforcement Learning, Fine-Tuning, Zero-Shot Learning, Continual Learning, Online Learning, Multi-Agent Learning, Supervised Learning, Instruction Tuning |
| Performance Highlights | None |
| Application Domains | General scientific literature review and knowledge synthesis, Artificial intelligence / machine learning research, Biomedicine / biology (protocol planning, hypothesis generation, drug discovery), Chemistry / materials science (hypothesis discovery, experimental planning, robotic chemistry), Physics (function discovery, gravitational physics benchmarks), Data science (code generation, data analysis, tabular reasoning), Cross-domain agentic research / autonomous scientific discovery platforms |
348. A Survey of Scientific Large Language Models: From Data Foundations to Agent Frontiers, Preprint (August 28, 2025)
| Category | Items |
|---|---|
| Datasets | Intern-S1 continual pre-training corpus (scientific subset), Galactica pre-training corpus, MedPaLM-2 training/eval datasets (medical-domain datasets / USMLE-style), ProteinChat dataset (protein prompt-answer triplets), LLaMA-Gene instruction corpus (DNA/protein tasks), HuatuoGPT-II corpus, NatureLM corpus (multidisciplinary biomedical + materials), MIMIC-CXR, PMC-OA / PMC-CaseReport / PMC figures datasets, ZINC, ChEMBL, USPTO reaction dataset, Materials Project, MatBench, UniProtKB / Swiss-Prot, PDB (Protein Data Bank), UniRef / UniRef50 / UniRef90 (protein sequence clusters), MD17 / ISO17 (molecular dynamics), Kepler mission data / TESS / ZTF mentioned, LIGO/Virgo GWTC catalogs, ERA5 / WeatherBench / WEATHER-5K, RS5M / GeoLLaVA-8K / GeoPixel / LHRS datasets (remote sensing corpora), MMSci / MaCBench (materials multimodal benchmarks), ScienceQA, MMLU-Pro, ResearchBench, HLE (Humanity’s Last Exam), SFE (Scientists’ First Exam) |
| Models | BERT, GPT, Transformer, Vision Transformer, CLIP, U-Net, Attention Mechanism, Self-Attention Network, Multi-Head Attention, Graph Neural Network, Message Passing Neural Network, Convolutional Neural Network |
| Tasks | Question Answering, Text Generation, Language Modeling, Image-to-Image Translation, Image Classification, Regression, Classification, Time Series Forecasting, Sequence-to-Sequence, Image Generation, Clustering, Feature Extraction, Anomaly Detection, Symbolic Regression, Molecular Property Prediction (mapped to Regression/Classification), Image Captioning, Information Retrieval, Hyperparameter Optimization / Architecture Search |
| Learning Methods | Transfer Learning, Pre-training, Fine-Tuning, Reinforcement Learning, Self-Supervised Learning, Continual Learning, Supervised Learning, Prompt Learning, Few-Shot Learning, Zero-Shot Learning, Contrastive Learning, Test-Time Learning, Representation Learning |
| Performance Highlights | accuracy: >85% (MedPaLM-2 on USMLE-style questions, as reported), benchmarks: state-of-the-art on PubMedQA and MedMCQA-dev (Galactica reported at release), sample_efficiency: Matches FNO performance with only 20 samples vs FNO needing 1024 samples, accuracy: 86.2% (AstroSage-LLaMA-3.1 on AstroMLab-1 benchmark, reported), accuracy: ≈98.6% (CSLLM on synthesizability prediction task), metastable_generation_rate: ≈49% materials generated metastable (CrystaLLM reported), HLE_performance: Closed-source models show steep drops on HLE (frontier scientific benchmark); Grok 4 reported 50.7% on HLE in paper, claimed: Intern-S1 ‘surpasses existing closed-source state-of-the-art models in professional tasks such as molecular synthesis, reaction condition prediction, and crystalline thermodynamic stability prediction’ (no numeric values reported in paper), win_or_draw_rate_vs_vicuna: 95% (Xiwu vs Vicuna-13B reported win-or-draw rate), code_generation_performance: Surpasses GPT-4 on certain HEP code generation tasks (qualitative claim), improvement: Test-time training improves chemical exploration but diminishing returns beyond long test-time durations (cited study results qualitative) |
| Application Domains | Physics, Chemistry, Materials Science, Life Sciences / Biology / Multi-omics, Healthcare / Medical Science, Astronomy, Earth Science / Remote Sensing / Climate, Agriculture, Neuroscience, Pharmacy / Drug Discovery |
347. High-power lithium-ion battery characterization dataset for stochastic battery modeling, Scientific Data (August 28, 2025)
| Category | Items |
|---|---|
| Datasets | High-power lithium-ion battery characterization dataset (this work) |
| Models | None |
| Tasks | None |
| Learning Methods | Ensemble Learning |
| Performance Highlights | None |
| Application Domains | High-power lithium-ion batteries, Electric vehicles (EVs), High-performance racing cars, Electric vertical take-off and landing (eVTOL) aircraft, Battery modeling and diagnostics (SoH/SOC/state estimation, stochastic modeling) |
346. Graph attention networks decode conductive network mechanism and accelerate design of polymer nanocomposites, npj Computational Materials (August 28, 2025)
| Category | Items |
|---|---|
| Datasets | hPF-MD simulated CNT/homopolymer dataset (CP1-CP8), Additional CNT variants (four other CNTs) (supplementary) |
| Models | Graph Neural Network, Graph Convolutional Network, Attention Mechanism, Multi-Head Attention |
| Tasks | Regression, Dimensionality Reduction, Clustering, Feature Extraction, Representation Learning |
| Learning Methods | Supervised Learning, Incremental Learning, Fine-Tuning, Pre-training, Backpropagation, Gradient Descent, Representation Learning, End-to-End Learning |
| Performance Highlights | CP1_RMSE_GAT: 0.00022, CP1_MAE_GAT: 0.00022, CP2_RMSE_GAT: 0.00025, CP2_MAE_GAT: 0.00025, CP3_RMSE_GAT: 0.00016, CP3_MAE_GAT: 0.00016, CP4_RMSE_GAT: 0.00023, CP4_MAE_GAT: 0.00023, CP5_RMSE_GAT: 0.00017, CP5_MAE_GAT: 0.00017, CP6_RMSE_GAT: 0.00012, CP6_MAE_GAT: 0.00012, CP7_RMSE_GAT: 9e-05, CP7_MAE_GAT: 9e-05, CP8_RMSE_GAT: 0.00021, CP8_MAE_GAT: 0.00021, aggregate_observation: prediction errors reported in Table 1 (RMSE and MAE) for GAT are approximately in the 1e-4 range per concentration, CP1_RMSE_GCN: 0.01762, CP1_MAE_GCN: 0.01762, CP2_RMSE_GCN: 0.01872, CP2_MAE_GCN: 0.01872, CP3_RMSE_GCN: 0.01208, CP3_MAE_GCN: 0.01208, CP4_RMSE_GCN: 0.01588, CP4_MAE_GCN: 0.01588, CP5_RMSE_GCN: 0.01586, CP5_MAE_GCN: 0.01586, CP6_RMSE_GCN: 0.01472, CP6_MAE_GCN: 0.01472, CP7_RMSE_GCN: 0.01224, CP7_MAE_GCN: 0.01224, CP8_RMSE_GCN: 0.01211, CP8_MAE_GCN: 0.01211, aggregate_observation: GCN baseline RMSE/MAE are on the order of 1e-2 per Table 1 |
| Application Domains | Conductive polymer nanocomposites (CPNs), Materials science, Predictive materials design, Wearable electronics / flexible electronics, Strain sensors and electronic skin, Soft robotics |
345. Deep generative models design mRNA sequences with enhanced translational capacity and stability, Science (August 28, 2025)
| Category | Items |
|---|---|
| Datasets | GEMORNA-CDS training dataset (natural protein sequences and corresponding CDSs), GEMORNA-UTR pre-training dataset (5’ and 3’ UTRs), GEMORNA-UTR fine-tuning datasets, Pred-5UTR training dataset (mean ribosome load labels), Pred-3UTR training dataset (stability labels), Bicknell et al. dataset (m1Ψ-modified exogenous mRNAs), Leppek et al. dataset (unmodified exogenous mRNAs), In-house m1Ψ-modified Fluc2P dataset, Experimental benchmarking datasets (in vitro / in vivo assays used for performance evaluation) |
| Models | Transformer, Seq2Seq, Encoder-Decoder, Bidirectional LSTM, Conditional Random Field, Recurrent Neural Network, Convolutional Neural Network |
| Tasks | Sequence-to-Sequence, Language Modeling, Data Generation, Regression, Synthetic Data Generation |
| Learning Methods | Unsupervised Learning, Pre-training, Fine-Tuning, Supervised Learning, Zero-Shot Learning |
| Performance Highlights | improvement_over_biLSTM-CRF_at_48h_Fluc: up to 20-fold, improvement_over_pGL4.11_at_48h_Fluc: 4.8-fold, 5prime_UTRs_similar_or_higher_than_BNT162b2: 5 GEMORNA 5’ UTRs exhibited similar or higher Fluc activities compared with BNT162b2 5’ UTR, group_level_improvement_over_BNT162b2_UTR_pairs: up to 7-fold increase in Fluc activity compared to the benchmark BNT162b2 UTRs, HEK293T_vs_HepG2_correlation: r^2 = 0.92, UTR_pair_cross_target_correlation: r^2 = 0.096, max_fold_increase_Fluc_vs_Benchmark-FL1_in_vitro: 41-fold, fold_increase_Fluc_vs_Benchmark-FL2_in_vitro: 8.2-fold, fold_increase_Fluc_in_HepG2_vs_Benchmark-FL2: 15.9-fold, antibody_titers_vs_BNT162b2_and_LinearDesign: GEMORNA-derived full-length mRNA induced higher antibody titers than both the BNT162b2 and the LinearDesign mRNAs at multiple time points (no numeric titer provided in text), NanoLuc_improvement_vs_benchmark: GEMORNA-designed mRNAs exhibited higher expression than a strong benchmark across 24-72 hours (no single fold-number provided in main text), EPO_in_vitro: 6 of 7 GEMORNA designs achieved enhanced EPO activities in vitro; selected designs showed longer durability, EPO_in_vivo_best_fold_increase: GMR-EPO-F7 achieved 15-fold increase in expression compared to the benchmark at 24 hours, circRNA_in_vitro_EPO_accumulated_expression_vs_benchmark: 13.8-fold (best GEMORNA design vs Chen et al. benchmark, Fig. 5F), circRNA_in_vivo_EPO_best_fold_increase_at_24h: 121-fold increase vs benchmark (Fig. 5H), circRNA_expression_longevity_ratio_best: 46.5% (144h/24h) vs benchmark 2.5% (Fig. 5G), NanoLuc_circRNA_long_durability: GMR-NL3 maintained high NanoLuc activity over 144 hours, and achieved a 3-fold increase at 72h vs 24h (Fig. 5C-E), training_dataset_size: 166,530 sequences, data_split: 0.7/0.15/0.15 (train/val/test), reported_performance: described as ‘high accuracy’ in selection of 5’ UTRs for fine-tuning (no numeric accuracy provided in text), training_dataset_size: 90,000 sequences, data_split: 0.7/0.15/0.15 (train/val/test), reported_performance: used to identify stable 3’ UTRs for fine-tuning (no numeric metric in main text), correlation_with_Bicknell_invitro_half-life: r = 0.92, correlation_with_Bicknell_invivio_expression: r = 0.96, relative_performance: GEMORNA CDSs up to 20-fold improvement over biLSTM-CRF at 48h Fluc |
| Application Domains | mRNA therapeutics and vaccines, synthetic biology (RNA design), protein replacement therapy (e.g., EPO), vaccine antigen design (COVID-19 spike protein), non-viral CAR-T cell therapy (CD19 CAR via circRNA), circular RNA (circRNA) therapeutics, gene expression optimization in mammalian cells |
344. One-shot design of functional protein binders with BindCraft, Nature (August 27, 2025)
| Category | Items |
|---|---|
| Datasets | PD-1 designs (this work), PD-L1 designs (this work), IFNAR2 designs (this work), CD45 designs (this work), CLDN1 (claudin 1) soluble-analogue designs (this work), BBF-14 designs (de novo beta-barrel target, this work), SAS-6 designs (this work), Der f7 designs (dust mite allergen; this work), Der f21 designs (dust mite allergen; this work), Bet v1 designs (birch allergen; this work), SpCas9 (CRISPR–Cas9) designs (this work), CbAgo (Clostridium butyricum Argonaute) designs (this work), HER2 and PD-L1 targeted AAV transduction screens (this work), PDB target inputs used for design (templates) |
| Models | Transformer, Message Passing Neural Network, Diffusion Model, Ensemble (multiple model weights), Rosetta (physics-based scoring) (NOT in provided model list) |
| Tasks | Structured Prediction, Data Generation, Binary Classification, Ranking, Regression |
| Learning Methods | Backpropagation, Stochastic Gradient Descent, Ensemble Learning, Pre-training, Representation Learning, Gradient Descent |
| Performance Highlights | in_silico_initial_pass_rate_range: 16.8%–62.7%, mpnnsol_filtered_pass_rate_range: 0.6%–65.9%, experimental_success_rate_range_per_target: 10%–100%, average_experimental_success_rate: 46.3%, mpnnsol_sequence_design_limit: Only two MPNNsol sequences per AF2 trajectory allowed to pass filters (to promote interface diversity), mpnnsol_contribution: Improved expression/stability in many designs (qualitative; pass rates reflected in filtered designs), generation_time_comparison: BindCraft yields similar success rates in terms of generation time across several targets and binder lengths compared with RFdiffusion, amino_acid_distribution_difference: RFdiffusion-generated designs underrepresent bulky amino acids at binder interface (qualitative), i_pTM_as_binary_predictor: AF2 i_pTM effectively discriminates on-target vs off-target interactions (qualitative/ROC-like behaviour reported); i_pTM does not correlate with interaction affinity, example: i_pTM used to rank designs; top designs showed experimental binding |
| Application Domains | Therapeutics (binder therapeutics, allergy neutralization), Biotechnology (AAV retargeting for gene delivery), Structural biology (de novo protein design and structural validation), Molecular biology (modulation of nucleases such as SpCas9 and CbAgo), Immunology (immune checkpoint receptor binders, allergen neutralization) |
343. Digital Twin for Chemical Science: a case study on water interactions on the Ag(111) surface, Nature Computational Science (August 27, 2025)
| Category | Items |
|---|---|
| Datasets | Ambient-pressure X-ray photoelectron spectroscopy (APXPS) spectra of Ag(111) interacting with H2O (Ag/H2O system) |
| Models | Gaussian Process, Hidden Markov Model, Markov Random Field |
| Tasks | Regression, Time Series Forecasting, Optimization, Simulation (mapped to ‘Time Series Forecasting’ and ‘Regression’ tasks) |
| Learning Methods | Supervised Learning, Stochastic Learning, Model-Based Learning |
| Performance Highlights | iterations: 1000, relative_error_cutoff: 0.5 (unitless), qualitative_accuracy: Improved with additional spectra; with more input spectra GP performance approaches basin hopping but was slower in convergence in reported tests, qualitative_match: Surface CRN showed slightly better match to experimental spectra than bulk CRN; surface CRN required more time to reach equilibrium, runtime_iterations: Not applicable (simulation-based forward solver); Gillespie stochastic runs used, qualitative: Bulk CRN outputs smooth concentration profiles; surface CRN outputs stochastic concentration profiles showing uncertainty due to Markov chain randomness |
| Application Domains | Chemical characterization / spectroscopy (APXPS), Surface science (metal surface — Ag(111)), Catalysis and electrocatalysis, Corrosion, Battery interfaces / electrode–electrolyte interfaces, Autonomous / on-the-fly experimental planning and decision support |
342. Target-aware 3D molecular generation based on guided equivariant diffusion, Nature Communications (August 25, 2025)
| Category | Items |
|---|---|
| Datasets | PDBbind (2020), CrossDocked (refined subset) |
| Models | Denoising Diffusion Probabilistic Model, Graph Neural Network, Multi-Layer Perceptron |
| Tasks | Graph Generation, Synthetic Data Generation, Optimization, Regression, Feature Extraction |
| Learning Methods | Supervised Learning, Representation Learning, Backpropagation, Stochastic Gradient Descent, End-to-End Learning |
| Performance Highlights | JS_C-C_2A_PDBbind: 0.1815, JS_all-atom_12A_PDBbind: 0.0486, JS_bonds_mean_PDBbind: 0.363 ± 0.15, JS_angles_mean_PDBbind: 0.211 ± 0.08, JS_dihedrals_mean_PDBbind: 0.361 ± 0.10, JS_bonds_mean_CrossDocked: 0.392 ± 0.18, JS_angles_mean_CrossDocked: 0.198 ± 0.05, JS_dihedrals_mean_CrossDocked: 0.423 ± 0.13, RMSD_median_approx: ~1.0 Å (DiffGui consistently achieves ~1 Å RMSD across scenarios), Vina_Score_mean_PDBbind: -6.700 ± 2.55, Vina_Min_mean_PDBbind: -7.655 ± 2.51, Vina_Dock_mean_PDBbind: -8.448 ± 2.24, QED_mean_PDBbind: 0.631 ± 0.21, SA_mean_PDBbind: 0.678 ± 0.15, LogP_mean_PDBbind: 1.977 ± 3.01, TPSA_mean_PDBbind: 100.49 ± 62.97, Validity_DiffGui-nolab: 0.9427 (higher validity noted), Ablation_effects: Removal of bond diffusion or property guidance leads to deteriorated JS divergences, Vina scores, and QED; removing both results in larger performance drop, LeadOptimization_examples: Generated candidates show Vina/QED/SA comparable or superior to reference ligands in case studies (e.g., PDBid 3l13, 6e23, DHODH experiments)., WetLab_IC50_RSK4: Compound 1 IC50 ≈ 215.0 nM; Compound 2 IC50 ≈ 111.1 nM, WetLab_IC50_DHODH: Compound 3 IC50 improved from 8.02 μM to 4.27 μM; Compound 4 IC50 improved from 32.20 nM to 10.45 nM |
| Application Domains | Structure-based drug design (SBDD), De novo drug design, Lead optimization (fragment-based design, scaffold hopping), Protein-ligand 3D molecular generation, Computational medicinal chemistry / virtual screening |
341. A versatile multimodal learning framework bridging multiscale knowledge for material design, npj Computational Materials (August 25, 2025)
| Category | Items |
|---|---|
| Datasets | Multimodal electrospun nanofibers dataset, Nanofiber-reinforced composite dataset |
| Models | Multi-Layer Perceptron, Convolutional Neural Network, ResNet, Transformer, Vision Transformer, Diffusion Model, Denoising Diffusion Probabilistic Model |
| Tasks | Regression, Information Retrieval, Image Generation, Optimization, Representation Learning, Multi-task Learning |
| Learning Methods | Self-Supervised Learning, Contrastive Learning, Pre-training, Fine-Tuning, Transfer Learning, Multi-Task Learning, Representation Learning, Classifier-Free Guidance |
| Performance Highlights | R2: MatMCL (conditions) and MatMCL (fusion) reported higher R2 vs conventional models without SGPT (exact numeric R2 values not reported in main text figures), RMSE: MatMCL (conditions) shows significant improvement (lower RMSE) vs conventional models without SGPT (exact numeric RMSE values not reported in main text figures), R2/RMSE: Transformer-style MatMCL evaluated and shows decreasing multimodal contrastive loss during training and competitive test performance; architecture choice has little impact on retrieval and both architectures evaluated for regression (exact numeric metrics not provided in text), Feature_correlations: Orientation seen: 0.968; Orientation unseen: 0.926; Diameter seen: 0.954; Diameter unseen: 0.947; Pore size seen: 0.882; Pore size unseen: 0.604, FID: FID heatmap reported showing much lower FID on diagonal (generated vs real for same condition); exact numeric FID per-condition values not listed in main text, RMSE: Multi-stage learning (MSL) achieves much lower prediction error on composite test set compared to models trained from random initialization; exact numeric RMSE values not provided in main text, Prediction accuracy: Predicted fracture strengths closely match measured values for both nanofibers and composites; prediction errors for all validated samples remained within a narrow range (exact values in Supplementary / figures), Top-k retrieval accuracy: MatMCL achieves significantly higher retrieval accuracy compared to random and a similarity-based baseline; exact top-1/top-3/top-5/top-10 numeric values are shown in Fig. 3 but not explicitly enumerated in main text, KNN on representations (structural feature prediction): Representations of MatMCL (fusion) achieve highest KNN regression performance for predicting structural features; MatMCL (conditions) next; original processing conditions worst (Fig. 2g). |
| Application Domains | Materials science, Electrospun nanofiber design and characterization, Nanofiber-reinforced composite design, Multimodal materials databases (cross-modal retrieval/generation), Inverse materials design / process optimization |
340. GraphVelo allows for accurate inference of multimodal velocities and molecular mechanisms for single cells, Nature Communications (August 22, 2025)
| Category | Items |
|---|---|
| Datasets | dyngen simulated datasets (linear, cyclic, bifurcating), Analytical 3D bifurcation system constrained to 2D manifold, FUCCI cell cycle dataset, Pancreatic endocrinogenesis dataset, Dentate gyrus dataset, Intestinal organoid dataset, Hematopoiesis dataset, Mouse erythroid maturation (mouse gastrulation -> erythroid lineage subset), Human bone marrow development dataset, Mouse coronal hemibrain spatial transcriptomics (binned; bin size 60), HCMV-infected monocyte-derived dendritic cells (moDCs) dataset, SARS-CoV-2 infected Calu-3 cells (Perturb-seq dataset subset), SHARE-seq mouse hair follicle dataset (multi-omics; transcriptome + ATAC), Developing human cortex 10x Multiome multi-omics dataset, A549 sci-fate metabolic-labeling cell cycle dataset |
| Models | Variational Autoencoder |
| Tasks | Time Series Forecasting, Regression, Clustering, Dimensionality Reduction, Feature Extraction, Causal Inference, Representation Learning, Clustering |
| Learning Methods | Unsupervised Learning, Representation Learning, Dimensionality Reduction, Gradient Descent, Representation Learning |
| Performance Highlights | cosine_similarity: GraphVelo preserves both direction and magnitude vs cosine kernel (figures show significant improvements), RMSE: GraphVelo shows lower RMSE vs cosine kernel in simulated tests (Fig.2b, f–h), accuracy: GraphVelo shows higher accuracy in sign prediction vs cosine kernel and random predictor (Fig.2f–h), CBC_score: GraphVelo achieved noticeably improved cross-boundary correctness (CBC) score against input velocity and other advanced methods (Supplementary Fig. 2–4), Spearman_pseudotime_vs_embryo_time: ρ = 0.831 (GraphVelo vector field-based pseudotime vs embryo time for erythroid lineage; Fig.3d), Spearman_pseudotime_vs_viral_RNA%: ρ = 0.980 (GraphVelo pseudotime vs % viral RNA in HCMV dataset; Fig.4b); scVelo baseline: ρ = 0.601 (p = 2.79E-143), Jacobian-based_regulatory_maps: Recovered sequential activation (Gata2 -> Gata1 -> Klf1) in erythropoiesis and identified viral factors inhibiting host pathways (HCMV analyses); in silico knockouts indicate UL123 as top viral factor reducing total viral RNA, in_silico_knockout_screen_samples: n = 1454 (virtual perturbation screen in HCMV; Fig.4j) |
| Application Domains | single-cell transcriptomics (developmental biology, cell differentiation), single-cell multi-omics (scRNA + scATAC; chromatin dynamics), spatial transcriptomics (mouse brain spatial datasets), host-pathogen interactions / infection dynamics (HCMV, SARS-CoV-2), simulated single-cell data for benchmarking (dyngen, analytic dynamical systems) |
339. Capturing short-range order in high-entropy alloys with machine learning potentials, npj Computational Materials (August 21, 2025)
| Category | Items |
|---|---|
| Datasets | TS-0, TS-1, TS-2, TS-3, TS-4, TS-5, TS-f (final training set), Test set (from ref. 23) |
| Models | Radial Basis Function Network |
| Tasks | Regression, Distribution Estimation, Clustering |
| Learning Methods | Supervised Learning, Ensemble Learning, Batch Learning |
| Performance Highlights | energy_RMSE_at_1800K_TS-0: 5.3 meV/atom, energy_RMSE_at_1800K_TS-3: 5.6 meV/atom, energy_RMSE_at_2684K_TS-0: 7.6 meV/atom, energy_RMSE_at_2684K_TS-3: 6.6 meV/atom, ε_SRO_relative_error_TS-2_vs_DFT: TS-2 reproduces DFT WC parameters within statistical accuracy (qualitative), ensemble_std_at_levmax_20_TS-2: 3%, ensemble_std_at_levmax_20_TS-0: 29%, ensemble_std_at_levmax_20_TS-1: 13%, pRDF_first_shell_peak_height_agreement_TS-3_vs_DFT: within ±4.9%, pRDF_first_shell_peak_location_discrepancy_Cr-Cr: -0.12 Å, ε_pRDF_relative_error_TS-3_vs_DFT: TS-3 better than TS-0 (specific numeric values shown in Fig. 2c), TS-4_failure_to_reproduce_γ_sf_and_ΔE: TS-4 (fcc-only) is not capable of reproducing γ_sf and ΔE to DFT accuracy (Fig. 3 shows mismatch), TS-5_success_reproduce_SRO_effects: TS-5 (includes hcp) captures correct SRO and its effects on γ_sf and ΔE (qualitative/within DFT statistical accuracy as shown in Fig. 3), melting_temperature_TS-f: 1661 K, experimental_melting_temperature: ~1690 K, note: TS-f melting temperature shows excellent agreement with experiment; improvement over EAM (EAM result not reliably parsed from main text formatting). |
| Application Domains | Atomistic simulations / computational materials science, High-entropy alloys (CrCoNi) and metallic alloys, Prediction of materials properties: stacking-fault energy, phase stability, melting temperature, Study of chemical short-range order (SRO) in crystal and liquid phases |
338. Machine learning-assisted Ru-N bond regulation for ammonia synthesis, Nature Communications (August 21, 2025)
| Category | Items |
|---|---|
| Datasets | 201 Ru-based binary intermetallics curated from ICSD, High-throughput DFT adsorption dataset (subset used as labels) |
| Models | XGBoost, Multi-Layer Perceptron, Gradient Boosting Tree, Support Vector Machine |
| Tasks | Regression, Ranking, Feature Selection, Feature Extraction |
| Learning Methods | Supervised Learning, Ensemble Learning |
| Performance Highlights | R2_EN_train: 0.98, R2_EN_test: 0.92, R2_EN2_train: 0.94, R2_EN2_test: 0.82 |
| Application Domains | Ammonia synthesis catalysis, Materials informatics / computational materials discovery, Catalyst screening and design (Ru-based intermetallic compounds), DFT-driven materials modeling combined with ML |
337. Revealing nanostructures in high-entropy alloys via machine-learning accelerated scalable Monte Carlo simulation, npj Computational Materials (August 20, 2025)
| Category | Items |
|---|---|
| Datasets | FeCoNiAlTi DFT dataset (this work), MoNbTaW DFT dataset (from ref. 33, used here), FeCoNiAlTi MC simulation configurations (this work), MoNbTaW MC simulation configurations (this work), Simulated APT specimen configurations (this work) |
| Models | Graph Neural Network, Multi-Layer Perceptron, Generalized Linear Model, Polynomial Model |
| Tasks | Regression, Clustering, Feature Extraction, Hyperparameter Optimization |
| Learning Methods | Supervised Learning, Backpropagation, Stochastic Gradient Descent |
| Performance Highlights | validation_RMSE: 0.1819 mRy/atom (~2.5 meV/atom), R^2: >0.995, validation_RMSE: 0.1643 mRy/atom, R^2: >0.995 |
| Application Domains | Computational materials science, Atomistic simulations of high-entropy alloys, Nanostructure evolution and nanoparticle morphology, Thermodynamic finite-temperature simulations (order-disorder transitions), Data-driven Monte Carlo simulations accelerated by machine-learned energy models, Comparison with experimental characterization (APT, TEM) |
336. Bayesian learning-assisted catalyst discovery for efficient iridium utilization in electrochemical water splitting, Science Advances (August 20, 2025)
| Category | Items |
|---|---|
| Datasets | DFT screening dataset of 66 binary oxides (OER overpotential), IrTiO2 Bayesian-optimization dataset (ΔGO − ΔGOH), IrTiO2−x oxygen-vacancy dataset (ΔGO − ΔGOH), Formation energy / stability dataset for IrxTi1−xO2 configurations, Materials Project rutile oxide unit cells, Zenodo repository (trained ML models & datasets) |
| Models | Gaussian Process, Radial Basis Function Network, Multi-Layer Perceptron |
| Tasks | Regression, Regression, Regression, Optimization, Experimental Design, Feature Extraction, Dimensionality Reduction |
| Learning Methods | Supervised Learning, Active Learning, Batch Learning, Feature Learning |
| Performance Highlights | MAE_ΔG(ΔGO−ΔGOH): 0.06 eV, MAE_formation_energy: 0.004 eV/atom, MAE_ΔG_IrTiO2−x: 0.10 eV, Predicted_mass_activity_enhancement: >40x (predicted at 12.5% Ir surface ratio with oxygen vacancies) |
| Application Domains | Electrocatalysis (Oxygen Evolution Reaction), Materials discovery / computational materials science, Catalyst design and synthesis, Hydrogen production / renewable energy (PEM water electrolysis) |
335. A unified pre-trained deep learning framework for cross-task reaction performance prediction and synthesis planning, Nature Machine Intelligence (August 19, 2025)
| Category | Items |
|---|---|
| Datasets | Pre-training dataset (13 million chemical reactions; 6.8 million high-quality), USPTO-50k, USPTO-full, USPTO-480k, USPTO-STEREO, Buchwald–Hartwig reaction dataset (dataset 1), Suzuki–Miyaura reaction dataset (dataset 2), Radical C–H functionalization dataset (dataset 3), Asymmetric thiol addition dataset (dataset 4), External validation dataset 1 (Ni-catalysed C–O coupling / NiCOlit), External validation dataset 2 (asymmetric hydrogenation of olefins), External validation dataset 3 (pallada-electrocatalysed C–H activation), Related datasets referenced in external validation splitting |
| Models | Graph Neural Network, Transformer, Graph Convolutional Network, Multi-Layer Perceptron, Seq2Seq, Random Forest, BERT, Graph Neural Network, Transformer, Graph Attention Network |
| Tasks | Regression, Classification, Sequence-to-Sequence, Clustering, Representation Learning |
| Learning Methods | Pre-training, Contrastive Learning, Fine-Tuning, Supervised Learning, Representation Learning, Contrastive Learning |
| Performance Highlights | dataset1R2: 0.971, dataset1_m.a.e._yield%: 2.98, dataset2R2: 0.876, dataset2_m.a.e._yield%: 6.30, dataset3R2: 0.992, dataset3_m.a.e.∆G_kcal/mol: 0.266, dataset4R2: 0.915, dataset4_m.a.e.∆∆G_kcal/mol: 0.134, component_combination_test_m.a.e.yield%: 10.12, external_dataset1m.a.e._yield%: 21.76, external_dataset1R2: 0.309, external_dataset1_binary_precision_high-yield: 0.793, external_dataset1_binary_accuracy_high-yield: 0.732, external_dataset2_R2: 0.832, external_dataset2_m.a.e.∆∆G_kcal/mol: 0.371, external_dataset3R2: 0.924, external_dataset3_m.a.e.∆∆G_kcal/mol: 0.211, USPTO-50k_top-1accuracy_retrosynthesis%: 51.0, USPTO-50k_top-3accuracy_retrosynthesis%: 69.0, USPTO-50k_top-5accuracy_retrosynthesis%: 74.2, USPTO-50k_top-10accuracy_retrosynthesis%: 79.2, USPTO-full_top-1accuracy_retrosynthesis%: 47.4, USPTO-full_top-3accuracy_retrosynthesis%: 63.0, USPTO-full_top-5accuracy_retrosynthesis%: 67.4, USPTO-full_top-10accuracy_retrosynthesis%: 71.6, USPTO-480k_top-1accuracy_forward%: 90.6, USPTO-480k_top-3accuracy_forward%: 94.3, USPTO-480k_top-5accuracy_forward%: 94.9, USPTO-480k_top-10accuracy_forward%: 95.5, USPTO-STEREO_top-1accuracy_forward%: 78.2, USPTO-STEREO_top-3accuracy_forward%: 85.1, USPTO-STEREO_top-5accuracy_forward%: 86.5, USPTO-STEREO_top-10accuracy_forward%: 87.8 |
| Application Domains | Organic synthesis, Chemical reaction prediction (reactivity, yield), Selectivity prediction (regioselectivity, enantioselectivity), Computer-aided synthesis planning (retrosynthesis and forward synthesis), Computational chemistry / catalysis, Data-driven reaction discovery and reaction space exploration |
334. Boosting the predictive power of protein representations with a corpus of text annotations, Nature Machine Intelligence (August 18, 2025)
| Category | Items |
|---|---|
| Datasets | Swiss-Prot (UniProtKB Swiss-Prot subset, 2023-02), Temporal test set (Swiss-Prot entries added 2023-02 to 2024-01), Binary localization, Subcellular localization, Fold, DAVIS, BindingDB, UniRef50 / BFD100 / UniParc (pretraining corpora referenced) |
| Models | Transformer, Encoder-Decoder, Seq2Seq, Attention Mechanism, Cross-Attention, Multi-Head Attention, Self-Attention Network, BERT, T5, GPT, Linear Model |
| Tasks | Representation Learning, Multi-class Classification, Multi-label Classification, Binary Classification, Sequence Labeling, Regression, Information Retrieval, Few-Shot Learning, Language Modeling |
| Learning Methods | Fine-Tuning, Pre-training, Self-Supervised Learning, Language Modeling, Few-Shot Learning, Contrastive Learning, Supervised Learning, Transfer Learning |
| Performance Highlights | average_improvement_over_base_family_%: 10.89, average_improvement_over_base_name_%: 12.53, average_improvement_over_base_domain_%: 10.55, average_improvement_over_base_binding_site_%: 8.49, average_improvement_over_base_active_site_%: 9.84, improvement_family_%: 15.10, improvement_name_%: 13.56, improvement_domain_%: 17.31, improvement_binding_site_%: 13.55, improvement_active_site_%: 12.41, improvement_over_base_ESM2-650M_%: 19.39, improvement_over_base_ESM1b_%: 7.47, improvement_over_base_ProtT5%: 11.89, average_improvement_GO-CC%: 7.08, average_improvement_GO-MF_%: 12.12, average_improvement_GO-BP_%: 5.84, Pearson_correlation_improvement: positive (exact values in Supplementary Table B; reported qualitatively as improved across models), tasks_outperforming_BLAST_count: 6of_9, tasks_with_comparable_performance: 3_of_9, one_shot_accuracy_PAIR_embeddings%: 87.1 ± 0.8, one_shot_accuracy_baselines_range_%: 67–77, low_resource_one_shot_average_accuracy_PAIR_%: ≈85 (P < 0.02 vs baseline) |
| Application Domains | Protein function prediction / protein annotation, Protein representation learning (bioinformatics), Enzyme function classification (EC prediction), Subcellular localization prediction, Protein fold / remote homology detection, Drug–target interaction (binding affinity prediction), Retrieval of annotated proteins (bioinformatics sequence search and retrieval) |
333. Steering towards safe self-driving laboratories, Nature Reviews Chemistry (August 18, 2025)
| Category | Items |
|---|---|
| Datasets | Syn-TODD, Vector-LabPics, TransProteus CGI dataset, Solution-based inorganic materials synthesis procedures dataset, ChemDataExtractor-generated datasets (examples: perovskite and dye-sensitized solar-cell device databases), HeinSight / HeinSight2.0 datasets (implied) |
| Models | Convolutional Neural Network, Transformer, GPT, Diffusion Model, Multi-Layer Perceptron |
| Tasks | Instance Segmentation, Depth Estimation, Object Localization, Image Classification, Anomaly Detection, Language Modeling, Text Generation, Named Entity Recognition, Experimental Design, Optimization, Sequence Labeling |
| Learning Methods | Active Learning, Reinforcement Learning, Imitation Learning, End-to-End Learning, Fine-Tuning, Pre-training, Multi-Agent Learning, Supervised Learning |
| Performance Highlights | success_rate: 71% |
| Application Domains | chemistry (autonomous chemical experimentation, synthesis, reaction optimization), materials science (materials discovery, thin-film materials, solid-state synthesis), robotics (laboratory robotic manipulation, mobile robots, motion planning), computer vision (laboratory perception, transparent object detection, liquid monitoring), natural language processing (protocol extraction, named entity recognition, LLM-based protocol conversion), autonomous vehicles (comparison and safety lessons drawn from AV domain), biological SDLs and space-based SDLs (discussed as cross-domain applicability) |
332. An automated framework for exploring and learning potential-energy surfaces, Nature Communications (August 18, 2025)
| Category | Items |
|---|---|
| Datasets | GAP-RSS dataset (silicon), GAP-RSS dataset (TiO2, Ti–O binary system), GAP-RSS dataset (SiO2 @ PBE and @SCAN), GAP-RSS dataset (liquid water / ice polymorphs) @ revPBE-D3(zero), GAP-RSS dataset (phase-change materials: Ge1Sb2Te4 and In3Sb1Te2), Materials Project relaxation trajectories (referenced as common dataset for foundational MLIPs) |
| Models | Gaussian Process, Graph Neural Network, Message Passing Neural Network, Feedforward Neural Network |
| Tasks | Regression, Data Generation, Feature Extraction |
| Learning Methods | Supervised Learning, Active Learning, Pre-training, Fine-Tuning, Transfer Learning, Incremental Learning |
| Performance Highlights | target_accuracy: 0.01 eV per atom (10 meV/atom), silicon_diamond_beta-tin: ≈0.01 eV/atom achieved with ≈500 DFT single-point evaluations, silicon_oS24: ≈0.01 eV/atom achieved within ‘a few thousand’ DFT single-point evaluations, RMSE_meV_per_atom_TiO2_Anatase (GAP-RSS TiO2-only): 0.1, RMSE_meV_per_atom_TiO2_Anatase (GAP-RSS Full Ti–O system): 0.7, RMSE_meV_per_atom_TiO2_Baddeleyite (TiO2-only): 1.1, RMSE_meV_per_atom_TiO2_Baddeleyite (Full Ti–O): 28, RMSE_meV_per_atom_TiO2_Brookite: 10 (TiO2-only) ; 8.2 (Full Ti–O), RMSE_meV_per_atom_TiO2_Columbite: 1.0 ; 0.9, RMSE_meV_per_atom_TiO2_Rutile: 0.2 ; 1.8, RMSE_meV_per_atom_TiO2-B: 24 ; 20, RMSE_meV_per_atom_Ti3O5 (TiO2-only trained model): 105 ; Full Ti–O: 19, notable_errors: Errors >1 eV/atom for some off-stoichiometry attempts when model trained only on TiO2 (reported as >1 eV/atom and not tabulated numerically)., ΔE_meV_per_atom_coesite (PBE DFT): 30 (DFT), 31 (GAP-RSS), ΔE_meV_per_atom_stishovite (PBE DFT): 186 (DFT), 185 (GAP-RSS), ΔE_meV_per_atom_α-cristobalite (PBE DFT): −7.9 (DFT) ; GAP-RSS −7.5 (PBE) — note PBE erroneously predicts cristobalite more stable than α-quartz, ΔE_meV_per_atom_moganite (PBE DFT): −0.4 (DFT) ; GAP-RSS −3.5 (PBE) — PBE inaccuracies noted, GAP@SCAN vs GAP@PBE: GAP@SCAN yields qualitatively correct sign for α-quartz vs α-cristobalite (ΔE>0), whereas GAP@PBE gives ΔE<0 (incorrect ordering)., liquid_water_hydrogen_bond_number_NequIP: ≈3.5 (average predicted by NequIP fit to GAP-RSS dataset), experimental_hydrogen_bond_range_reference: 3.48 to 3.84 (literature range cited), ice_structures_energy_prediction: NequIP shows ‘much improved predictive accuracy’ versus GAP when predicting energies of 54 ice structures (qualitative improvement; numeric errors not tabulated in main text), Ge1Sb2Te4_atomic_environments_reduction: 46% fewer atomic environments used in GAP-RSS-derived model compared to hand-crafted training dataset of ref.26 (which had 49,056 environments) — implies ≈26,500 atomic environments (approximate), crystallisation_simulation_time_GAP: crystallisation simulation essentially completed after 350 ps (GAP-driven MD), qualitative_performance: RDFs and ring statistics show encouraging agreement with AIMD references (Fig.5c–f), but exact numeric RMSEs not provided in main text |
| Application Domains | Computational materials science, Inorganic solids (silicon, TiO2, Ti–O binaries, SiO2 polymorphs), Condensed-phase molecular systems (liquid water, ice polymorphs), Phase-change memory materials (Ge1Sb2Te4, In3Sb1Te2), High-throughput ML-driven materials exploration / potential-energy surface exploration |
330. Accelerated design of gold nanoparticles with enhanced plasmonic performance, Science Advances (August 15, 2025)
| Category | Items |
|---|---|
| Datasets | Electrodynamics simulation results (BEM) for AuBP parameter sweeps, Experimental characterization dataset of gold bipyramids (AuBPs) |
| Models | Gaussian Process, Gaussian Process |
| Tasks | Optimization, Hyperparameter Optimization, Experimental Design |
| Learning Methods | Bayesian Optimization, Multi-Objective Learning, Batch / Quasi-Random Sampling (initialization technique) |
| Performance Highlights | TMax_K_at_λ_exc_1050nm: 679, Optimal_dimensions_length_nm: 137, Optimal_dimensions_width_nm: 27, Absorption_to_extinction_ratio: ≈0.80, EFSERS_at_λ_exc_1050nm: 9.3e7, EFSERS_local_maxima_950nm_values: ∼8.4e7 and ∼7.9e7, Optimal_dimensions_length_nm: 140, Optimal_dimensions_width_nm: 29, Optimal_analyte_distance_nm: 2, PLEF_max_at_λems_950nm: 5965, Quantum_yield_enhancement_max_at_λexc_850nm: 0.8, Optimal_dimensions_length_nm: 140, Optimal_dimensions_width_nm: 35, Optimal_emitter_distance_nm: 2, Rabi_splitting_range_meV: 150-200, Example_ΩR_meV_at_λems_650nm: 195, Optimal_dimensions_length_nm: 66, Optimal_dimensions_width_nm: 31, Optimal_excitonic_layer_thickness_nm: 5, ΔE94_max: ≈4.79, Optimal_dimensions_length_nm: 58, Optimal_dimensions_width_nm: 40, Optimal_interparticle_gap_nm: 2, ΔH_max_percent: ≈86, Pareto_optimization_example_objectives: maximize PLEF while minimizing δT (λ = 950 nm), Human_in_loop_steps_equivalent: BO pipeline accelerates optimization ~600× relative to canonical human-in-the-loop OFAT (estimated OFAT ~31,000 steps to search 3D parameter space) |
| Application Domains | Materials Science, Nanophotonics, Plasmonics, Sensing / Colorimetric Sensing, Surface-Enhanced Raman Spectroscopy (SERS) / Molecular Sensing, Biomedicine (photothermal therapy, bioimaging), Optoelectronics / Quantum technologies (strong plexcitonic coupling) |
329. Chem3DLLM: 3D Multimodal Large Language Models for Chemistry, Preprint (August 14, 2025)
| Category | Items |
|---|---|
| Datasets | QM9, Cross-Docked (CrossDocked) dataset |
| Models | Transformer, Multi-Layer Perceptron, Graph Neural Network, Normalizing Flow, Diffusion Model, Convolutional Neural Network, Transformer |
| Tasks | Molecular Conformation Generation, Structure-Based Drug Design |
| Learning Methods | Supervised Learning, Fine-Tuning, Reinforcement Learning, Policy Gradient, Pre-training, Multi-Task Learning, Representation Learning |
| Performance Highlights | Atom Stability (%): 99.45, Mol Stability (%): 95.00, Valid (%): 100.00, Unique (%): 100.00, Vina Score Avg. (lower better): -7.03, Vina Med. (median): -7.15, Vina Min. (best min after optimization, joint training): -12.30, Vina Score Avg. (joint multi-task Chem3DLLM†): -7.21, Vina Score Avg. (w/o RLSF / SFT only): -7.03, Vina Min. (w/o RLSF / SFT only): -12.20, Vina Score Avg. (w/o RCMT): -1.82, Vina Min. (w/o RCMT): -4.70, Atom Stability (%): 85.0, Mol Stability (%): 4.9, Valid (%): 40.2, Unique (%): 39.4, Atom Stability (%): 95.7, Mol Stability (%): 68.1, Valid (%): 85.5, Unique (%): 80.3, GDM Atom Stability (%): 97.0, GDM Mol Stability (%): 63.2, GDM-AUG Atom Stability (%): 97.6, GDM-AUG Mol Stability (%): 71.6, EDM Atom Stability (%): 98.7, EDM Mol Stability (%): 82.0, EDM-Bridge Atom Stability (%): 98.8, EDM-Bridge Mol Stability (%): 84.6, GeoLDM Atom Stability (%): 98.9, GeoLDM Mol Stability (%): 89.4, Atom Stability (%): 98.25, Mol Stability (%): 86.87, Valid (%): 100.0, Unique (%): 100.0, Reference Vina Avg.: -6.36, Reference Vina Med.: -6.46, AR Vina Avg.: -5.75, AR Vina Med.: -5.64, Pocket2Mol Vina Avg.: -5.14, Pocket2Mol Vina Med.: -4.70, FLAG Vina Avg.: 16.48, FLAG Vina Med.: 4.53, TargetDiff Vina Avg.: -5.47, TargetDiff Vina Med.: -6.30, Decomp-R Vina Avg.: -5.19, Decomp-R Vina Med.: -5.27, Decomp-O Vina Avg.: -5.67, Decomp-O Vina Med.: -6.04, MolCRAFT Vina Avg.: -6.59, MolCRAFT Vina Med.: -7.04 |
| Application Domains | Computational chemistry, Drug discovery / Structure-based drug design (SBDD), Molecular modeling and conformation generation, Materials science (3D molecular structure modeling), Protein-ligand interaction modeling |
327. SAGERank: inductive learning of protein–protein interaction from antibody–antigen recognition, Chemical Science (August 12, 2025)
| Category | Items |
|---|---|
| Datasets | Antibody–Antigen docking dataset (authors’ training set), Cognate antibody–antigen pairing dataset (shuffled pairings), TCR–pMHC dataset (STCRDab), Small protein–protein docking decoy set (authors), Expanded protein–protein docking test set (authors), DC dataset (biological vs crystal interfaces), Cancer target epitope benchmark (IEDB-annotated), Molecular glue ternary complex experiments (FKBP12–rapamycin–FRAP), Nanobody–protein recognition test set (DeepConformer / AlphaFold3 comparison), Reference / prior antibody–antigen benchmarks (cited) |
| Models | Graph Neural Network, Graph Convolutional Network |
| Tasks | Ranking, Binary Classification, Structured Prediction |
| Learning Methods | Supervised Learning, End-to-End Learning, Inductive Learning, Representation Learning |
| Performance Highlights | training_set_size_complexes: 287 complexes, total_decoys: 455,420, comparison: outperforms ZRANK, PISA, FoldX and Rosetta on Ab–Ag docking decoy set (see Fig. 2B), AUC_test_set: 0.82, average_score_positive: 0.57, average_score_negative: 0.20, F1_max_at_threshold_0.3: 0.74, confusion_matrix_test_counts: negatives: 9813/11307 correctly classified; positives: 3685/4642 correctly classified, ROCAUC_residue_level_GCa: 0.6467, PRAUC_residue_level_GCa: 0.6739, small_PPI_set_performance: SAGERank competitive with or slightly outperforming PISA on some aspects (see Fig. 2B), expanded_PPI_test_set: on 80 complexes (62,220 structures) SAGERank significantly surpassed success rates of three other scoring methods and was on par with PISA, binding_site_prediction_proteins: 8 out of 10 protein cases correct (80% accuracy), epitope_prediction_antigens: 3 out of 5 antigen cases correct (60% accuracy), DC_dataset_accuracy_SAGERank: 80%, DC_dataset_accuracy_PISA: 79%, DC_dataset_accuracy_PRODIGY: 74%, DC_dataset_accuracy_Deep-Rank: 86%, molecular_glue_positive_total_native_in_top10_SAGERank: 30 native ternary complexes identified in top-10 selections (across positive conformations aggregated as ‘All’ in Table 4), molecular_glue_positive_total_native_in_top10_Pisa: 10, molecular_glue_negative_total_native_in_top10_SAGERank: 2, molecular_glue_negative_total_native_in_top10_Pisa: 6, mean_min_iRMSD_sagerank: 6.62 Å, mean_min_iRMSD_af3_score: 6.88 Å, median_min_iRMSD_sagerank: 5.75 Å, median_min_iRMSD_af3_score: 5.32 Å, near_native_rate_iRMSD<=2.0_sagerank: 25.6%, near_native_rate_iRMSD<=2.0_af3_score: 27.9%, correlation_between_methods: strong positive correlation (r > 0.7) between iRMSD values from both methods |
| Application Domains | antibody–antigen recognition and antibody design, structural immunology (epitope prediction, TCR–pMHC specificity), protein–protein interaction prediction and docking, molecular glue / ternary complex screening (protein + small molecule), cancer antigen epitope identification, general computational structural biology / bioinformatics |
326. Probing the limitations of multimodal language models for chemistry and materials research, Nature Computational Science (August 11, 2025)
| Category | Items |
|---|---|
| Datasets | MaCBench (v1.0.0) |
| Models | Transformer, Vision Transformer, GPT, BERT |
| Tasks | Classification, Multi-class Classification, Regression, Image Classification, Feature Extraction, Sequence-to-Sequence, Text Generation, Binary Classification |
| Learning Methods | Prompt Learning, Fine-Tuning, In-Context Learning, Pre-training |
| Performance Highlights | equipment_identification_accuracy: 0.77, table_composition_extraction_accuracy: 0.53, hand_drawn_to_SMILES_accuracy: 0.8, isomer_relationship_naming_accuracy: 0.24, stereochemistry_assignment_accuracy: 0.24, baseline_accuracy: 0.22, crystal_system_assignment_accuracy: 0.55, space_group_assignment_accuracy: 0.45, atomic_species_counting_accuracy: 0.85, capacity_values_interpretation_accuracy: 0.59, Henry_constants_comparison_accuracy: 0.83, XRD_amorphous_vs_crystalline_accuracy: 0.69, AFM_interpretation_accuracy: 0.24, MS_NMR_interpretation_accuracy: 0.35, XRD_highest_peak_identification_accuracy: 0.74, XRD_relative_intensity_ranking_accuracy: 0.28, performance_dependency_on_internet_presence: positive_correlation (visualized in Fig. 5) |
| Application Domains | chemistry (organic chemistry, spectroscopy, NMR, mass spectrometry), materials science (crystallography, MOF isotherms, electronic structure, AFM), laboratory experiment understanding and safety assessment, in silico experiments and materials characterization, scientific literature information extraction and data curation |
325. Unconditional latent diffusion models memorize patient imaging data, Nature Biomedical Engineering (August 11, 2025)
| Category | Items |
|---|---|
| Datasets | PCCTA (in-house photon-counting coronary CT angiography sub-volumes), MRNet, fastMRI, X-ray (ChestX-ray8 subset) |
| Models | Denoising Diffusion Probabilistic Model, Diffusion Model, Variational Autoencoder, Generative Adversarial Network, Transformer, Autoencoder |
| Tasks | Image Generation, Synthetic Data Generation, Novelty Detection, Data Augmentation |
| Learning Methods | Self-Supervised Learning, Contrastive Learning, Adversarial Training, Variational Inference, Generative Learning, Representation Learning |
| Performance Highlights | aggregate_memorization_pct: approx. 37.2% (abstract reported overall across datasets), aggregate_synthetic_copies_pct: approx. 68.7% (abstract reported overall across datasets), PCCTA_Nmem_pct: 43.8%, PCCTA_Ncopies_pct: 91.7%, MRNet_Nmem_pct: 40.2%, MRNet_Ncopies_pct: 76.1%, fastMRI_Nmem_pct: 24.8%, fastMRI_Ncopies_pct: 37.3%, PCCTA_Nmem_pct: 40.5%, PCCTA_Ncopies_pct: 83.1%, MRNet_Nmem_pct: 48.2%, MRNet_Ncopies_pct: 87.4%, fastMRI_Nmem_pct: 30.8%, fastMRI_Ncopies_pct: 51.0%, Xray_Nmem_pct: 32.6% (MONAI-2D), Xray_Ncopies_pct: 54.5% (MONAI-2D), VQVAE-Trans_PCCTA_Nmem_pct: 49.6%, VQVAE-Trans_MRNet_Nmem_pct: 58.3%, VQVAE-Trans_fastMRI_Nmem_pct: 40.2%, VQVAE-Trans_PCCTA_Ncopies_pct: 66.1%, VQVAE-Trans_MRNet_Ncopies_pct: 83.3%, VQVAE-Trans_fastMRI_Ncopies_pct: 57.5%, CCE-GAN_overall: Detected some copies but generally unable to synthesize realistic samples in 3D (many detected copies shared little global info), proj-GAN_2D: Synthesized reasonable quality images in 2D but contained no copies (only very small number of false positives), PCCTA_MedDiff_sensitivity: 97.6%, PCCTA_MedDiff_specificity: 93.1%, PCCTA_MONAI_sensitivity: 88.4%, PCCTA_MONAI_specificity: 94.7%, MRNet_MedDiff_sensitivity: 95.4%, MRNet_MedDiff_specificity: 90.6%, MRNet_MONAI_sensitivity: 100%, MRNet_MONAI_specificity: 85.5%, fastMRI_MONAI_sensitivity: 80.6%, fastMRI_MONAI_specificity: 94.3%, Xray_MONAI-2D_sensitivity: 83.3%, Xray_MONAI-2D_specificity: 94.2%, PCCTA_MedDiffAug_Nmem_pct: 40.1%, PCCTA_MONAIAug_Nmem_pct: 36.0%, PCCTA_MedDiffAug_Ncopies_pct: 72.7%, PCCTA_MONAIAug_Ncopies_pct: 76.3%, MRNet_MedDiffAug_Nmem_pct: 27.7%, MRNet_MONAIAug_Nmem_pct: 27.1%, MRNet_MedDiffAug_Ncopies_pct: 36.0%, MRNet_MONAIAug_Ncopies_pct: 61.5%, fastMRI_MedDiffAug_Nmem_pct: 8.7%, fastMRI_MONAIAug_Nmem_pct: 6.3%, fastMRI_MedDiffAug_Ncopies_pct: 9.3%, fastMRI_MONAIAug_Ncopies_pct: 6.4%, Xray_MONAIAug_Nmem_pct: 5.6%, Xray_MONAIAug_Ncopies_pct: 7.3%, architecture_params_MONAI_2D: 25m (small), 171m (medium), 270m (large), architecture_params_MONAI_3D: 68m (small), 191m (medium), 442m (large), general_observation: small architecture had lower memorization across datasets (except PCCTA); medium and large had higher but similar memorization |
| Application Domains | Medical imaging (MRI, CT, chest X-ray), Synthetic data generation / open-data sharing in healthcare, Patient privacy and re-identification risk assessment, Data augmentation and dataset expansion for downstream AI tasks, Evaluation of generative model memorization in 2D and 3D medical images |
324. Observation of dendrite formation at Li metal-electrolyte interface by a machine-learning enhanced constant potential framework, Nature Communications (August 11, 2025)
| Category | Items |
|---|---|
| Datasets | superlattice model (used for cell relaxation), Li/[EC + LiPF6] interfacial model (initial training dataset), single-interface model (used in configurational sampling), double-interface model (pair of counter electrodes) (used in configurational sampling and production MD), training and testing dataset (DFT-labeled; full dataset archive) |
| Models | Multi-Layer Perceptron, Feedforward Neural Network |
| Tasks | Regression, Data Generation |
| Learning Methods | Supervised Learning, Active Learning, Stochastic Gradient Descent, Representation Learning |
| Performance Highlights | full_DP_energy_RMSE_test: 3.04e-3 eV/atom, full_DP_force_RMSE_test: 1.54e-1 eV/Å, DP-QEq_ConstQ_energy_RMSE_test: 1.31e-3 eV/atom, DP-QEq_ConstQ_force_RMSE_test: 1.63e-1 eV/Å, DP-QEq_ConstP_energy_RMSE_test: 1.00e-2 eV/atom, DP-QEq_ConstP_force_RMSE_test: 2.27e-1 eV/Å, Li_atoms_force_RMSE_in_dendrite_regions: 0.07–0.09 eV/Å |
| Application Domains | Li metal batteries (Li metal anodes, solid electrolyte interphase), Electrochemical interface modeling, Atomistic simulations of dendrite nucleation, All-solid-state batteries (suggested broader application), Electrocatalytic surface corrosion modeling (suggested broader application) |
323. Designing Pb-Free High-Entropy Relaxor Ferroelectrics with Machine Learning Assistance for High Energy Storage, Journal of the American Chemical Society (August 06, 2025)
| Category | Items |
|---|---|
| Datasets | dataset of 141 titanate-based compositions |
| Models | Random Forest |
| Tasks | Regression, Feature Selection, Dimensionality Reduction, Hyperparameter Optimization, Feature Extraction |
| Learning Methods | Supervised Learning, Ensemble Learning |
| Performance Highlights | test_R2: 0.81, MAE: 0.7 J cm^-3, experimental_Wrec_of_ML-designed_composition: 17.2 J cm^-3, efficiency_eta: 87%, breakdown_strength_EB: 79 kV mm^-1 |
| Application Domains | Materials Science, Dielectric Energy Storage, Relaxor Ferroelectrics, High-Entropy Materials, Pb-free ceramic capacitors, Pulsed-power electronic systems |
321. Quantifying large language model usage in scientific papers, Nature Human Behaviour (August 04, 2025)
| Category | Items |
|---|---|
| Datasets | arXiv, bioRxiv, Nature portfolio (15 journals), Validation set (pre-ChatGPT) |
| Models | GPT, Transformer, Language Modeling |
| Tasks | Text Summarization, Text Generation, Distribution Estimation, Binary Classification, Text Classification, Information Retrieval, Feature Extraction, Language Modeling |
| Learning Methods | Maximum Likelihood Estimation, Prompt Learning, Zero-Shot Learning, Fine-Tuning, Embedding Learning, Adversarial Training, Contrastive Learning, Feature Extraction |
| Performance Highlights | population_level_prediction_error: < 3.5%, computer_science_abstracts_alpha_Sept_2024: 22.5% (bootstrapped 95% CI (21.7%, 23.3%)), computer_science_introductions_alpha_Sept_2024: 19.6% (bootstrapped 95% CI (19.2%, 20.0%)), electrical_engineering_abstracts_alpha_Sept_2024: 18.0% (bootstrapped 95% CI (16.7%, 19.3%)), electrical_engineering_introductions_alpha_Sept_2024: 18.4% (bootstrapped 95% CI (17.8%, 19.0%)), mathematics_abstracts_alpha_Sept_2024: 7.7% (bootstrapped 95% CI (7.1%, 8.3%)), nature_portfolio_abstracts_alpha_Sept_2024: 8.9% (bootstrapped 95% CI (8.2%, 9.6%)), validation_set_size_sentences_per_ground_truth_alpha: n = 30,000 sentences; validation range alpha = 0% to 25% in 5% increments, abstracts_more_similar_alpha_Sept_2024: 23.0% (bootstrapped 95% CI (22.3%, 23.7%)), abstracts_less_similar_alpha_Sept_2024: 18.7% (bootstrapped 95% CI (18.0%, 19.4%)), robustness_to_proofreading_estimated_increase: approx. +1% in estimated alpha after LLM ‘proofreading’ (minor edits), validation_role: Used to generate realistic LLM-produced training data which contributed to validation error < 3.5% (see above) |
| Application Domains | scientific publishing / academic writing, computer science research papers (arXiv), electrical engineering and systems science (arXiv), mathematics (arXiv), physics and statistics (arXiv), biology (bioRxiv), journal articles in Nature portfolio (multidisciplinary journals) |
320. Modeling protein conformational ensembles by guiding AlphaFold2 with Double Electron Electron Resonance (DEER) distance distributions, Nature Communications (August 02, 2025)
| Category | Items |
|---|---|
| Datasets | OpenFold training subset (fine-tuning set), OpenFold larger training set, PDBe-KB validation dataset, Benchmark set of 29 targets (59 conformations), PfMATE experimental DEER distance distributions, LmrP experimental DEER distance distributions, Pgp (ABCB1) experimental DEER distance distributions, Simulated DEER datasets (Kazmier method) - Simulation 1, Simulated DEER datasets (Kazmier method) - Simulation 2, Zenodo repository of training targets and benchmarking results |
| Models | Transformer, Attention Mechanism |
| Tasks | Structured Prediction, Clustering, Dimensionality Reduction |
| Learning Methods | Fine-Tuning, Pre-training, Transfer Learning, Representation Learning |
| Performance Highlights | PfMATE_unconstrained_Neff=5median_RMSD_to_OF: 0.87 Å, PfMATE_unconstrained_Neff=5_median_RMSD_to_IF: 5.19 Å, PfMATE_Experiment1_Neff=5_median_RMSD_to_IF: 2.11 Å, PfMATE_Experiment1_Neff=5_median_RMSD_to_OF: 3.53 Å, PfMATE_Experiment2_Neff=5_median_RMSD_to_IF: 1.32 Å, PfMATE_Experiment2_Neff=5_median_RMSD_to_OF: 4.71 Å, PfMATE_Simulation1_Neff=5_median_RMSD_to_IF: 1.19 Å, PfMATE_Simulation2_Neff=5_median_RMSD_to_IF: 1.22 Å, PfMATE_TM-score_Experiment1_full_MSA%predictions_TM>0.9to_IF: 93%, LmrP_unconstrained_Neff=5_median_RMSD_to_IF: 5.56 Å (initial unconstrained set), LmrP_Experiment1_Neff=5_median_RMSD_to_IF: 1.52 Å, LmrP_Experiment2_Neff=5_median_RMSD_to_IF: 1.19 Å, LmrP_Simulation1_Neff=5_median_RMSD_to_IF: 1.28 Å, LmrP_Simulation2_2constraints#models_RMSD<3Å: 58 out of 100 models, Pgp_unconstrained_median_RMSD_to_IF_narrow: 3.41 Å, Pgp_Experiment1ADP-Vi_median_RMSD_to_OF_references: 3.25 Å and 3.30 Å, Pgp_Simulation2_fullset_54pairs_success_rate_TM>=0.9_to_7ZK7: 100% (all predictions transitioned), Pgp_Simulation2_random_8_constraints#models_switched_to_target_7ZK7_TM>=0.9: 293 out of 500 (58.6%), Benchmark_29_targets_constrained_average_RMSD_to_target: 3.11 Å (average constrained predictions), Benchmark_29_targets_constrained_average_TM-score: 0.91 (average constrained predictions), Benchmark_paired_t-test_RMSD_improvement: p = 1.38E-05, Benchmark_success_rate_over_29_targets: >70% for all 29 targets; 20 targets achieved 100%, EMD_correlation_with_TM-scores: Lower EMD distances consistently correspond to higher TM-scores and better match to target conformations (examples: LmrP, PfMATE, Pgp cluster centroids RMSD/TM reported), LmrP_cluster_centroids_RMSD_to_OF: 0.81 Å (yellow centroid), LmrP_blue_centroid_RMSD_to_IF: 1.73 Å, TM-score = 0.96, PfMATE_blue_centroid_RMSD_to_IF: 2.03 Å, TM-score = 0.93, Pgp_PC1_cluster_series_RMSD_range_to_IF: RMSD decreased from 5.38 Å to 2.93 Å with TM-score increasing from 0.84 to 0.95 |
| Application Domains | Protein structure prediction, Structural biology, Protein conformational dynamics, Membrane transporter biology, Biophysical spectroscopy (DEER/EPR) integration with ML, Model-guided experimental design (spin-label pair optimization), Computational structural biophysics |
319. Navigating protein landscapes with a machine-learned transferable coarse-grained model, Nature Chemistry (August 2025)
| Category | Items |
|---|---|
| Datasets | CATH domains training set (50 protein domains), Dimer mono-/dipeptide dataset (~1,200 / 1,245 dimers), Decoy/augmented frames (noisy frames), Test proteins (unseen) — set of small peptides and proteins with PDB codes, PUMA–MCL-1 and PUMA–Ubiquitin systems (case studies), Ubiquitin mutational benchmark (experimental ΔΔG from Went & Jackson 2005) |
| Models | Graph Neural Network, Graph Neural Network |
| Tasks | Distribution Estimation, Clustering, Dimensionality Reduction, Regression, Feature Extraction, Clustering |
| Learning Methods | Supervised Learning, Representation Learning |
| Performance Highlights | qualitative_match_of_free_energy_landscapes: CG free energy landscapes reproduce folded/unfolded/metastable basins comparable to all-atom reference for many test proteins, extrapolation_sequence_similarity_range: test proteins have low sequence similarity to training set (16–40% for many test proteins; Table 1 lists specific percents), Pearson_correlation_coefficient_r: 0.63, MAE: 1.25 kcal mol^-1, homeodomain_mean_RMSD_to_crystal: ~0.5 nm, homeodomain_fraction_native_contacts_Q: ~0.75, PUMA_induced_folding_r.m.s.d.: ~2.5 Å (0.25 nm threshold referenced), landscape_visualization: free energy landscapes plotted on TICA coordinates show CGSchNet captures many metastable states similar to all-atom reference (qualitative), comparison_to_other_CG_models: CGSchNet explores much of the all-atom free energy landscape; AWSEM/UNRES/Martini often stabilize a single metastable state, transferability: model extrapolates to proteins with low sequence similarity (examples in Table 1), and stabilizes folded states of larger proteins that were withheld from training, speedup: orders of magnitude faster than all-atom MD (qualitative; Supplementary Table 9 referenced) |
| Application Domains | protein folding and conformational dynamics, molecular dynamics / computational chemistry, structural biology, protein stability and mutational effect prediction, protein–peptide binding and folding-upon-binding studies, coarse-grained modeling and force-field development |
318. Data-driven de novo design of super-adhesive hydrogels, Nature (August 2025)
| Category | Items |
|---|---|
| Datasets | NCBI adhesive protein dataset (adhesive proteins), Resilin protein dataset, DM-driven hydrogel dataset (initial experimental dataset), Expanded hydrogel dataset (training + validation after ML rounds) |
| Models | Gaussian Process, Random Forest, Gradient Boosting Tree, XGBoost, Support Vector Machine, Linear Model |
| Tasks | Regression, Optimization, Feature Extraction, Dimensionality Reduction, Experimental Design, Feature Selection, Synthetic Data Generation |
| Learning Methods | Supervised Learning, Ensemble Learning, Batch Learning |
| Performance Highlights | model_selection: GP and RFR achieved the lowest RMSE among nine models (exact RMSE values not specified in main text), role_in_BO: GP_KB and RFR-GP were top-performing SMBO methods; GP_KB identified high-Fa formulations, model_selection: RFR was runner-up to GP in RMSE; selected as an effective base model, SMBO_best: RFR-GP produced the highest Fa overall among SMBO methods; warm-start variant RFR-GP* exhibited the highest Fa among all models, usage: GBM used as one of EI maximizers in batched SMBO (RFR-GBM) but RFR-GP and GP_KB outperformed other combinations, benchmarking: XGBoost (XGB) included among benchmarked non-linear models; not reported as top performer vs GP/RFR, benchmarking: Support vector regression (SVR) tested among non-linear models but did not achieve lowest RMSE, benchmarking: Lasso and Ridge linear regressions used as linear baselines; inferior to non-linear models (GP, RFR) in RMSE, experimental_dataset_growth: Initial dataset 180 → after round 1 added 109 validated points (total 289) → rounds 2 and 3 added 27 and 25 (final 341), hydrogel_performance: ML-driven hydrogels achieved Fa exceeding 1 MPa (R1-max); best DM-driven hydrogel (G-max) had Fa = 147 kPa; among 180 DM-driven gels 16 had Fa > 100 kPa and 83 had Fa > 46 kPa, durability: R1-max maintained robust adhesion over 200 attachment–detachment cycles; R1-max sustained 1-kg shear load for over 1 year |
| Application Domains | Soft materials design, Adhesive hydrogels / biomaterials, Biomedical engineering (implantation, wound sealing), Marine environments / deep-sea exploration, Marine farming and seawater applications, Polymer chemistry and materials discovery |
317. Electron-density-informed effective and reliable de novo molecular design and optimization with ED2Mol, Nature Machine Intelligence (August 2025)
| Category | Items |
|---|---|
| Datasets | DUD-E, ASB-E (AlloSteric Benchmark—Enhanced), PDB-derived pocket–ligand training set (processed subset), GP dataset (Growth-Point dataset), TA dataset (Torsion-Angle dataset), ZINC subset (drug-like molecules), Real-world target case sets (FGFR3, CDC42, GCK, GPRC5A) |
| Models | Variational Autoencoder, Graph Neural Network, Convolutional Neural Network, Multi-Layer Perceptron, Diffusion Model, Graph Neural Network (baseline mappings) |
| Tasks | Graph Generation, Optimization, Binary Classification, Multi-class Classification, Generative Learning, Representation Learning, Regression, Ranking |
| Learning Methods | Supervised Learning, Generative Learning, Representation Learning, Fine-Tuning, Classification, Adversarial Training |
| Performance Highlights | GP_dataset_size: 2,986,278 samples (from 94,519 molecules), TA_dataset_size: 17,978,368 samples (from 4,497,302 molecules), Generation_success_rate_DUD-E: 67.3%, Generation_success_rate_ASB-E: 68.9%, PB-valid (PoseBusters) percentage: 97.3 ± 0.4%, Occupancy_ranking_score: 28.2 (ED2Mol) > TargetDiff 26.2 > Pocket2Mol 25.6 > FLAG 15.0 > ResGen 7.9 > GraphBP 4.5, Occupancy_coverage: 84% (ED2Mol) vs 74% (TargetDiff), Reliable_success_rate: 34.4 ± 0.5% (ED2Mol) vs TargetDiff 17.1 ± 1.0% and Pocket2Mol 15.0 ± 2.0%, Re-docking_stability_reference: reference co-crystal ligands recovered 72% (RMSD < 2 Å), Re-docking_stability_notes: ED2Mol showed the highest fidelity; ‘12.7% more stable poses than the second-best model, Pocket2Mol, on DUD-E (34.8% total)’ and ‘15.9% better stability than the second-best model, TargetDiff, on ASB-E (29.0% total)’., PB-valid (PoseBusters) percentages: TargetDiff 58.1 ± 0.5% (PB-valid), Reliable_success_rate (reported baseline): TargetDiff 17.1 ± 1.0% (reliable success rate), PB-valid percentages: Pocket2Mol 63.1 ± 0.4%; FLAG 59.4 ± 1.4%; ResGen 34.1 ± 3.9%; GraphBP 32.2 ± 1.2%, Generation_success_rate_notes: ResGen and GraphBP produce poor binding molecules scored higher than zero (qualitative remark)., Reliable_success_rate_notes: Pocket2Mol 15.0 ± 2.0%; FLAG 6.7 ± 0.1%; ResGen 4.5 ± 1.1%, FGFR3: F4 KD = 599.0 μM (SPR); F42 KD = 61.4 μM (SPR), 9.8-fold improvement, Brr2 (hit optimization): Recovered inhibitor 9; docking RMSD = 0.56 Å (ED2Mol-generated vs re-docked lead), PPARγ (fragment growth): Recovered activator 3 with RMSD = 0.95 Å, CDC42: C1 IC50 = 47.58 ± 3.71 μM (in vivo BRET assay); C11 IC50 = 111.63 ± 0.90 μM; C1 KD = 5.35 μM (SPR), GCK: G1 EC50 = 290 nM (6.1-fold over G0); G11 EC50 = 150 nM (11.9-fold improvement), GPRC5A: A4 EC50 = 8.94 ± 2.30 μM (PRESTO-Tango assay); ED2Mol vs AlphaFold top-ranked holo structure RMSD = 1.48 Å |
| Application Domains | Structure-based drug discovery, De novo molecular design, Hit identification and lead optimization, Protein–ligand binding / computational structural biology, Allosteric modulator discovery, Cheminformatics / fragment-based molecular generation, Experimental validation (biophysical and cellular assays) |
316. Kolmogorov–Arnold graph neural networks for molecular property prediction, Nature Machine Intelligence (August 2025)
| Category | Items |
|---|---|
| Datasets | BACE, BBBP, ClinTox, SIDER, Tox21, HIV, MUV |
| Models | Graph Neural Network, Graph Convolutional Network, Graph Attention Network, Multi-Layer Perceptron |
| Tasks | Graph Classification, Binary Classification, Multi-label Classification, Regression |
| Learning Methods | Supervised Learning, End-to-End Learning, Backpropagation, Pre-training, Contrastive Learning |
| Performance Highlights | BACE (ROC-AUC): 0.890(0.014), BBBP (ROC-AUC): 0.787(0.014), ClinTox (ROC-AUC): 0.992(0.005), SIDER (ROC-AUC): 0.842(0.001), Tox21 (ROC-AUC): 0.799(0.005), HIV (ROC-AUC): 0.821(0.005), MUV (ROC-AUC): 0.834(0.009), BACE (ROC-AUC): 0.884(0.004), BBBP (ROC-AUC): 0.785(0.021), ClinTox (ROC-AUC): 0.991(0.005), SIDER (ROC-AUC): 0.847(0.002), Tox21 (ROC-AUC): 0.800(0.006), HIV (ROC-AUC): 0.823(0.002), MUV (ROC-AUC): 0.834(0.010), BACE (ROC-AUC): 0.835(0.014), BBBP (ROC-AUC): 0.735(0.011), ClinTox (ROC-AUC): 0.979(0.004), SIDER (ROC-AUC): 0.834(0.001), Tox21 (ROC-AUC): 0.747(0.006), HIV (ROC-AUC): 0.762(0.005), MUV (ROC-AUC): 0.741(0.006), BACE (ROC-AUC): 0.834(0.012), BBBP (ROC-AUC): 0.707(0.007), ClinTox (ROC-AUC): 0.983(0.006), SIDER (ROC-AUC): 0.836(0.002), Tox21 (ROC-AUC): 0.751(0.007), HIV (ROC-AUC): 0.761(0.003), MUV (ROC-AUC): 0.784(0.019), BACE (ROC-AUC): 0.890(0.014), BBBP (ROC-AUC): 0.787(0.014), ClinTox (ROC-AUC): 0.992(0.005), SIDER (ROC-AUC): 0.842(0.001), Tox21 (ROC-AUC): 0.799(0.005), HIV (ROC-AUC): 0.821(0.005), MUV (ROC-AUC): 0.834(0.009), BACE (ROC-AUC): 0.853(0.027), BBBP (ROC-AUC): 0.654(0.009), ClinTox (ROC-AUC): 0.981(0.004), SIDER (ROC-AUC): 0.832(0.006), Tox21 (ROC-AUC): 0.715(0.004), HIV (ROC-AUC): 0.804(0.010), MUV (ROC-AUC): 0.787(0.012), BACE (ROC-AUC): 0.771(0.012), BBBP (ROC-AUC): 0.723(0.008), ClinTox (ROC-AUC): 0.973(0.006), SIDER (ROC-AUC): 0.824(0.003), Tox21 (ROC-AUC): 0.724(0.005), HIV (ROC-AUC): 0.753(0.007), MUV (ROC-AUC): 0.638(0.008), BACE (ROC-AUC): 0.884(0.004), BBBP (ROC-AUC): 0.785(0.021), ClinTox (ROC-AUC): 0.991(0.005), SIDER (ROC-AUC): 0.847(0.002), Tox21 (ROC-AUC): 0.800(0.006), HIV (ROC-AUC): 0.823(0.002), MUV (ROC-AUC): 0.834(0.010), BACE (ROC-AUC): 0.8319(0.007), BBBP (ROC-AUC): 0.708(0.005), ClinTox (ROC-AUC): 0.983(0.003), SIDER (ROC-AUC): 0.836(0.001), Tox21 (ROC-AUC): 0.753(0.004), HIV (ROC-AUC): 0.818(0.006), MUV (ROC-AUC): 0.797(0.011), BACE (ROC-AUC): 0.808(0.009), BBBP (ROC-AUC): 0.657(0.004), ClinTox (ROC-AUC): 0.948(0.003), SIDER (ROC-AUC): 0.825(0.004), Tox21 (ROC-AUC): 0.731(0.012), HIV (ROC-AUC): 0.744(0.018), MUV (ROC-AUC): 0.792(0.013), Qualitative comparison: Fourier-based KAN demonstrates superior approximation capability compared to standard two-layer MLP across six representative functions (see Supplementary Fig. 1). |
| Application Domains | Molecular property prediction, Drug discovery, Computational chemistry, Biophysics, Physiology, Geometric deep learning for non-Euclidean data, General molecular data modelling |
315. An actor–critic algorithm to maximize the power delivered from direct methanol fuel cells, Nature Energy (August 2025)
| Category | Items |
|---|---|
| Datasets | Three-electrode chronoamperometry (CA) current–time trajectories (experimental), DMFC device long-term operation measurements (MEA experiments), Augmented dataset (synthetic adjacency-based augmentation) |
| Models | Convolutional Neural Network, Multi-Layer Perceptron, Feedforward Neural Network |
| Tasks | Control, Policy Learning, Decision Making, Regression, Data Augmentation, Optimization, Decision Making |
| Learning Methods | Reinforcement Learning, Policy Gradient, Temporal Difference Learning, Supervised Learning, Stochastic Gradient Descent, Gradient Descent |
| Performance Highlights | validation_MAE_mW: 0.011, Pearson_r: 0.969, alternate_plot_MAE_mW: 0.013, alternate_plot_r: 0.965, training_epochs_to_converge: 300, 4h_produced_power_mW_alphaFC: 0.284 ± 0.013, 4h_multiplier_vs_Co-Pt-Ru/NC_constant: 2.15×, 4h_multiplier_vs_PtRu/C_constant: 4.64×, 12h_time_averaged_power_vs_constant_percent: 153%, 12h_increase_vs_switching_strategy_percent: 30.4%, 12h_increase_vs_constant_strategies_percent: 185.2% and 486.1% (context-specific comparisons), 90h_average_power_multiplier_vs_constant: 4.86× (≈486%), long_term_stable_hours: >250 hours, actor_action_selection_time_CPU_s: ≈0.3, comparison_with_GA: GBO consistently achieves superior predicted produced power across batch sizes and run-time constraints (fig. 3c), runtime_tradeoff_note: Gradient tracking takes roughly three times more computations (justifying GA comparison), PID_power_fraction_of_alphaFC: ≈50%, MPC_note: MPC reliability degraded when simulation inaccurate; not numerically quantified here |
| Application Domains | Electrochemistry / Fuel cells (Direct Methanol Fuel Cells, DMFCs), Experimental device control / Real-world control systems, Energy devices: maximizing power delivery and prolonging catalyst life, Edge artificial intelligence for laboratory/device controllers, Potential generalization domains mentioned: battery formation/charging protocols, electrodeposition, temperature/fluid flow control in reactors |
313. Deep learning for property prediction of natural fiber polymer composites, Scientific Reports (July 30, 2025)
| Category | Items |
|---|---|
| Datasets | This study experimental natural-fiber composite dataset (augmented), Li et al. microstructure dataset (stochastic microstructures), MPOB degradable plastics dataset (Bakar et al.), Volgin synthetic polyimide dataset + experimental PIs, Gurnani et al. polymer dataset, polyBERT pretraining dataset and downstream dataset, Jung et al. optical property dataset, Aldeghi et al. polymer dataset (wD-MPNN), Xue et al. CFRP-wrapped RC columns dataset, Zhu et al. hydrogel SAW dataset, Bradford et al. SPE experimental dataset (ChemArr), QM9 and other referenced datasets |
| Models | Deep Neural Network, Multi-Layer Perceptron, Convolutional Neural Network, 3D Convolutional Neural Network, Graph Neural Network, Graph Convolutional Network, Message Passing Neural Network, Graph Attention Network, Long Short-Term Memory, Transformer, Random Forest, Gradient Boosting Tree, Linear Model, Deep Residual Convolutional Neural Network |
| Tasks | Regression, Regression, Sequence-to-Sequence, Regression, Regression, Representation Learning / Feature Extraction, Hyperparameter Optimization, Multitask Learning |
| Learning Methods | Supervised Learning, Transfer Learning, Pre-training, Fine-Tuning, Multi-Task Learning, Data Augmentation, Ensemble Learning, Hyperparameter Optimization, Feature Selection, AutoML, Representation Learning |
| Performance Highlights | Tensile Strength R2: 0.89 ± 0.01, Tensile Strength MAE: 2.1 ± 0.2 MPa, Young’s Modulus R2: 0.87 ± 0.02, Young’s Modulus MAE: 105 ± 7 MPa, Elongation at Break R2: 0.83 ± 0.02, Elongation at Break MAE: 1.3 ± 0.1 %, Impact Strength R2: 0.85 ± 0.02, Impact Strength MAE: 0.35 ± 0.03 kJ/m2, Tensile Strength R2: 0.88 ± 0.01, Tensile Strength MAE: 2.3 ± 0.2 MPa, Young’s Modulus R2: 0.84 ± 0.02, Young’s Modulus MAE: 115 ± 8 MPa, Elongation at Break R2: 0.81 ± 0.02, Elongation at Break MAE: 1.5 ± 0.1 %, Impact Strength R2: 0.83 ± 0.02, Impact Strength MAE: 0.4 ± 0.03 kJ/m2, Tensile Strength R2: 0.85 ± 0.02, Tensile Strength MAE: 2.7 ± 0.3 MPa, Young’s Modulus R2: 0.82 ± 0.03, Young’s Modulus MAE: 130 ± 10 MPa, Elongation at Break R2: 0.78 ± 0.03, Elongation at Break MAE: 1.8 ± 0.2 %, Impact Strength R2: 0.80 ± 0.03, Impact Strength MAE: 0.5 ± 0.05 kJ/m2, Tensile Strength R2: 0.72 ± 0.03, Tensile Strength MAE: 4.5 ± 0.4 MPa, Young’s Modulus R2: 0.65 ± 0.04, Young’s Modulus MAE: 210 ± 15 MPa, Elongation at Break R2: 0.60 ± 0.05, Elongation at Break MAE: 3.2 ± 0.3 %, Impact Strength R2: 0.68 ± 0.04, Impact Strength MAE: 0.9 ± 0.1 kJ/m2, Longitudinal modulus R2: 0.991, Transverse modulus R2: 0.969, In-plane shear modulus R2: 0.984, Major Poisson’s ratio R2: 0.903, Out-of-plane shear modulus R2: 0.955, Tg MAE: 20 K, Stress–strain curve prediction speed: seconds vs days for simulations, accuracy: high (qualitative; reported as fast, accurate predictions), Outperformance vs baselines: combined model outperformed both graph-only and sequence-only baselines (no single-number metric provided), λmax prediction improvement: reduced systematic errors for certain classes (qualitative); no single-number metric provided in text, Relative performance: outperformed Chemprop and XGBoost in both absolute error and ranking accuracy (no numeric values provided), Improved accuracy: significant improvement over unweighted MPNNs (no numeric metric provided) |
| Application Domains | Natural-fiber polymer composites (mechanical property prediction), Polymer informatics (polymer property prediction and fingerprinting), Soft materials and hydrogels (mechanical response prediction), Solid polymer electrolytes (ionic conductivity prediction for batteries), Molecular design for solar cells and optical properties (λmax prediction), Geopolymer concrete compressive strength prediction, Coarse-grained polymer modeling and accelerated molecular simulation, Structural engineering (CFRP-wrapped RC columns, lateral confinement coefficient) |
312. Accelerating primer design for amplicon sequencing using large language model-powered agents, Nature Biomedical Engineering (July 30, 2025)
| Category | Items |
|---|---|
| Datasets | SARS-CoV-2 synthetic RNA standards (Wuhan-01), Human genomic DNA standard NA12878, Mycobacterium tuberculosis (MTB) DNA standards and MTB culture isolates, Plasmid pools for enzyme mutant sequencing (Luc, KODm, Cid1, TdT), Curated microbial reference database (PrimeGen), ClinVar, OMIM, COSMIC, UniProt, CARD and WHO clinical-annotation datasets, SARS-CoV-2 variant file (derived) |
| Models | GPT, Transformer, Vision Transformer |
| Tasks | Information Retrieval, Optimization, Text Generation, Anomaly Detection, Classification, Embedding Learning, Pre-training, Sequence-to-Sequence |
| Learning Methods | Fine-Tuning, Prompt Learning, Pre-training, Representation Learning, Embedding Learning, Supervised Learning, In-Context Learning |
| Performance Highlights | sequence_search_success_rate_%: GPT-4o: 86; GPT-4: 84; Qwen-Max: 89; Qwen2.5-72B-Instruct: 88; GPT-3.5: 70; GLM-4-plus: 85, panel_optimization_reduction_in_loss_relative_per_method: LLM optimizer stabilizes loss comparable to stochastic greedy in 12-plex; in 78-plex LLM optimizer outperforms AdaLead and GA after ~750 iterations (no absolute loss value provided), code_modification_accuracy_%: GPT-4o: 100; GPT-4: 100; Qwen-Max: 100; Qwen2.5-72B-Instruct: 100; GPT-3.5: 88.75; GLM-4-plus: 85.2, Qwen2VL-7B_average_accuracy: 0.87, Qwen2VL-7B_slot-layout_accuracy: 0.79, Qwen2VL-7B_pipette-side_precision: 0.94, Qwen2VL-7B_well-plate-side_precision: 0.88, Qwen2VL-7B_BLEU: 0.20, Qwen2VL-7B_ROUGE-L: 0.50, Qwen2VL-7B_GPT4-Score: 4.25, retrieval_top_k: top-5 candidate code blocks retrieved via embedding similarity before final selection by GPT-4o |
| Application Domains | Biomedical research, Targeted next-generation sequencing (tNGS) and amplicon sequencing, Viral genomics (SARS-CoV-2 sequencing and variant surveillance), Clinical genetics (expanded carrier screening), Microbial pathogen detection and drug-resistance mutation detection (Mycobacterium tuberculosis), Protein engineering and directed evolution (plasmid mutant sequencing), Laboratory automation / self-driving laboratories / liquid-handling robotics, AI-driven protocol generation and code automation, Vision-based laboratory anomaly detection |
310. Geographic-style maps with a local novelty distance help navigate in the materials space, Scientific Reports (July 29, 2025)
| Category | Items |
|---|---|
| Datasets | Inorganic Crystal Structure Database (ICSD), Materials Project (MP), Cambridge Structural Database (CSD), Crystallography Open Database (COD), Berkeley A-lab dataset (A-lab crystals), GNoME training snapshot (subset of MP) |
| Models | Graph Neural Network |
| Tasks | Novelty Detection, Information Retrieval, Novelty Detection, Synthetic Data Generation |
| Learning Methods | Generative Learning, Representation Learning |
| Performance Highlights | training_dataset_size: 384,938 crystals (Materials Project 2021 snapshot), attempts_synthesized: 58 (attempted by A-lab), reported_materials_produced: 43, successes: 36, partial_successes: 7, duplicates_found_in_MP: 42 of the 43 A-lab crystals were found to already exist in the Materials Project (pre-dating the GNoME snapshot) |
| Application Domains | Materials science, Crystallography, Inorganic materials discovery, Autonomous/self-driving laboratories (automated synthesis), Database curation and integrity (duplicate detection) |
309. Biomimetic Intelligent Thermal Management Materials: From Nature-Inspired Design to Machine-Learning-Driven Discovery, Advanced Materials (July 29, 2025)
| Category | Items |
|---|---|
| Datasets | 119-compound DFT/phonon dataset, 911 datapoints from 25 studies (nanomaterial-enhanced PCM dataset), Molecular dynamics (MD) heat-flux dataset (training data for GAN), Polymer repeat-unit / MD-augmented polymer dataset (for CNN), 260 MD simulation results (for XGBoost seawater-evaporation model), Inverse-design / metasurface spectral response dataset (FDTD-simulated spectra), SETC performance dataset (solar absorption / storage metrics) |
| Models | Random Forest, Support Vector Machine, Gaussian Process, Multi-Layer Perceptron, Convolutional Neural Network, Generative Adversarial Network, Conditional GAN, Diffusion Model, Graph Neural Network, Long Short-Term Memory, Bidirectional LSTM, XGBoost, Gradient Boosting Tree, Feedforward Neural Network, Gaussian Mixture Model, Convolutional Neural Network (metasurface forward model) |
| Tasks | Regression, Time Series Forecasting, Optimization, Synthetic Data Generation, Ranking, Clustering, Feature Extraction, Image-to-Image (interpreted as structure→spectrum mapping / inverse design) |
| Learning Methods | Supervised Learning, Unsupervised Learning, Reinforcement Learning, Transfer Learning, Active Learning, Generative Learning, Adversarial Training, Evolutionary Learning, Pre-training, Fine-Tuning |
| Performance Highlights | dataset_size: 119 compounds, features: 57 features from crystal structure and composition, R2: 0.93, thermal_conductivity_Al: ~7 W m^-1 K^-1, thermal_conductivity_Cu: 13–14 W m^-1 K^-1, SEA_enhancement: 6–36x (initial SEA enhancement), recoverability_SEA_improvement: 0.2–1.5x, predicted_cooling_theoretical: 8.5 °C, experimental_cooling: 8.3 °C, reflectance: >95%, training_MD_samples: 260 MD results, candidate_materials_screened: 38,142 2D materials, validation_metrics: low mean absolute error and high coefficient of determination (reported qualitatively) |
| Application Domains | Electronics (device thermal management, semiconductor cooling), Aerospace (thermal protection and management), Biomedical (wearable thermal textiles, personal thermal management), Buildings (energy-efficient facades, smart windows, passive radiative cooling), Robotics (biomimetic robot thermal management), Photovoltaics (PV-leaf transpiration cooling, PV temperature reduction), Desalination / water harvesting (solar-driven atmospheric water harvesting, evaporation-based cooling), Energy storage (phase-change materials, solar-thermal storage), Metasurfaces / photonics (spectral regulation, infrared stealth) |
308. Atomistic Generative Diffusion for Materials Modeling, Preprint (July 24, 2025)
| Category | Items |
|---|---|
| Datasets | Quantum Cluster Database (QCD), Computational 2D Materials Database (C2DB) |
| Models | Denoising Diffusion Probabilistic Model, Graph Neural Network, Variational Autoencoder |
| Tasks | Data Generation, Synthetic Data Generation, Graph Generation |
| Learning Methods | Unsupervised Learning, Self-Supervised Learning, Representation Learning |
| Performance Highlights | Precision-Recall AUC (QCD GM clusters model): AUC value shown on PR curve in Fig.3(b) (higher than the larger ΔE<200 meV model); exact numeric AUCs are reported in the figure panels., Precision-Recall AUC (QCD ΔE<200 meV model): Lower AUC / lower recall compared to GM clusters model (value shown in Fig.3(b))., Precision-Recall AUC (C2DB model): PR curve and area under curve values shown in Fig.5(b) (figure labels show multiple baseline and model AUC values; model exhibits strong precision and recall)., Symmetry accuracy vs guidance scale: Symmetry accuracy peaks at moderate guidance scales (w ≈ 0.5–0.75) as shown in Fig.6(a); increasing guidance beyond w=1 degrades symmetry preservation., Qualitative plausibility of interpolated bimetallic clusters: t-SNE visualizations (Fig.4) show interpolated Pt–Cu and Pd–Ag clusters spanning configuration space between mono-metallic endpoints; sampled structures ‘appear visually similar to known bimetallic motifs’., Baseline PR curves (synthetic perturbations): Multiple synthetic baselines generated by adding Gaussian noise (σ varied) and subsampling (e.g., 100% and 50% coverage) produce PR curves shown in Fig.3(b) and Fig.5(b); these baselines are used as reference anchors. |
| Application Domains | Materials discovery, Nanoclusters / nanomaterials, Two-dimensional materials, Catalysis (mentioned as application domain), Energy storage materials, Crystal structure prediction (cited as potential application / future work), Inverse materials design |
307. Decoding nature’s grammar with DNA language models, Proceedings of the National Academy of Sciences (July 22, 2025)
| Category | Items |
|---|---|
| Datasets | 16 plant genomes (pretraining set), Maize genomes / maize variant calls (transfer evaluation), Arabidopsis annotated sites (benchmarks: transcription initiation and termination sites), Splice donor and acceptor site annotations (benchmarks), Site frequency spectrum / population variant frequency data, Sweet corn causal mutation (well-studied causal mutation), GERP++ constraint scores / alignment-based constraint datasets |
| Models | BERT, Transformer, Autoencoder |
| Tasks | Language Modeling, Sequence Labeling, Ranking, Anomaly Detection, Transfer Learning |
| Learning Methods | Self-Supervised Learning, Pre-training, Transfer Learning, Backpropagation, Cross-Entropy, Generative Learning, Unsupervised Learning |
| Performance Highlights | loss_metric: cross-entropy (perplexity used for evaluation), comparison: PlantCaduceus (state-space-based language model) performs as well as or better than prior alignment-based approaches; new SOTA baseline claimed (no numeric metrics reported in commentary) |
| Application Domains | Plant genomics, Variant effect prediction / prioritization, Annotation of noncoding genomic regions, Comparative genomics / evolutionary constraint analysis, Maize genetics (transfer application), Arabidopsis functional site annotation |
306. Uni-Electrolyte: An Artificial Intelligence Platform for Designing Electrolyte Molecules for Rechargeable Batteries, Angewandte Chemie (July 21, 2025)
| Category | Items |
|---|---|
| Datasets | DFT and MD database (electrolyte dataset), Embedded electrolyte database (EMolCurator), LiBE dataset (entire LiBE electrolyte database), USPTO chemical reaction database (pretraining data), QM9 (pretraining reference for Uni-Mol baseline), Reaxys reactions subset (augmentation) |
| Models | Graph Neural Network, Message Passing Neural Network, Diffusion Model, Denoising Diffusion Probabilistic Model, Encoder-Decoder, Ensemble (EMol-QSPR) |
| Tasks | Regression, Data Generation, Information Retrieval, Structured Prediction, Graph Generation |
| Learning Methods | Supervised Learning, Pre-training, Fine-Tuning, Ensemble Learning, Representation Learning, Transfer Learning |
| Performance Highlights | LEFTNet_OOD_MAE_dielectric_constant: 3.27, LEFTNet_OOD_MAE_viscosity_mPa_s: 12.97, G2GT_relative_error_vs_UniMol: 0.97, UniMol_relative_error_baseline: 1.0, G2GT_OOD_MAE_dielectric_constant: 3.31, G2GT_OOD_MAE_viscosity_mPa_s: 13.28, EMol-QSPR_relative_error_vs_baseline: 0.94, EMol-QSPR_OOD_MAE_dielectric_constant: 3.17, EMol-QSPR_OOD_MAE_viscosity_mPa_s: 12.83, improvement_over_G2GT_percent: 3, improvement_over_LEFTNet_percent: 4.4, qualitative_performance: EDM outperformed cG-Schnet in targeted HOMO–LUMO generation; successfully generated molecules including DME in sparse region, cG-LEFTNet_vs_cG-Schnet_Task2: comparable, cG-LEFTNet_vs_cG-Schnet_Task3: superior, G2GT_Top1_accuracy_one-step_retrosynthesis: 0.529, Askcos_Top1_accuracy_one-step_retrosynthesis: 0.452, G2GT-Askcos_number_of_retrosynthetic_routes_found: 17, ASKCOS_number_of_routes_found: 8, example_Gibbs_free_energy_ring_opening_eV: -4.98 |
| Application Domains | Rechargeable batteries (lithium-ion and lithium metal batteries), Electrolyte molecular design, Electrochemistry and interfacial (SEI) chemistry, Computational chemistry / materials discovery, Retrosynthetic planning for chemical synthesis |
305. AutoMAT: A Hierarchical Framework for Autonomous Alloy Discovery, Preprint (July 21, 2025)
| Category | Items |
|---|---|
| Datasets | Materials Properties Handbook: Titanium Alloys (handbook chapters parsed by LLM), TCHEA7 thermodynamic database (used via Thermo-Calc), Simulated candidate composition pools (AutoMAT-generated), Experimental dataset: as-cast titanium alloys (this work), Experimental dataset: as-cast HEAs (Al-Co-Cr-Fe-Ni) (this work) |
| Models | GPT, Transformer |
| Tasks | Information Retrieval, Synthetic Data Generation, Regression, Optimization, Ranking |
| Learning Methods | Pre-training, Model-Based Learning |
| Performance Highlights | latency: minutes, cost: less than US$1 (for the ideation query via GPT-4o API), throughput_examples: LLM completed alloy system identification, handbook analysis, and suggested candidate (Ti-185) within minutes, predicted_final_density: 4.355 g/cm3, predicted_final_yield_strength: 927.08 MPa, evaluation_rate: >1,000 compositions per day (multi-threaded execution), candidate_pool_reduction_Ti_case: from >43,000 potential compositions to 3,161 for detailed evaluation, time_reduction_Ti_case: reduced manual CALPHAD effort equivalent from ~2 years (100 compositions/day) to under a week, experimental_final_density: 4.32 g/cm3 (measured), experimental_final_yield_strength: 829 MPa (measured), density_reduction_vs_reference: 8.1% lower density vs reference Ti-185, specific_strength: 202 × 10^3 Pa·m^3/kg (reported high specific strength), predicted_HEA_final_yield_strength: 906.64 MPa (CALPHAD-predicted), predicted_HEA_improvement_pct: 70.4% predicted improvement over initial HEA candidate (532.11 MPa → 906.64 MPa), experimental_HEA_yield_strength_initial: 305 MPa (measured baseline), experimental_HEA_yield_strength_optimized: 397 MPa (measured) — reported up to 28.2% improvement, density_change_HEA: experimental density decreased from 7.33 g/cm3 to 7.17 g/cm3 |
| Application Domains | Materials science / alloy discovery, Metallurgy (titanium alloys, high-entropy alloys), Aerospace structural materials, Automotive structural materials, Biomedical (general potential — ideation layer applicability mentioned) |
304. DiffuMeta: Algebraic Language Models for Inverse Design of Metamaterials via Diffusion Transformers, Preprint (July 21, 2025)
| Category | Items |
|---|---|
| Datasets | Implicit-equation shell metamaterials dataset (D = {(Ψ(n), σ(n), C(n)) : n = 1 … N}), Evaluation sampling set (unconditional sampling), Fabricated experimental samples |
| Models | Denoising Diffusion Probabilistic Model, Transformer, Attention Mechanism, Self-Attention Network, Multi-Head Attention |
| Tasks | Synthetic Data Generation, Data Generation, Optimization, Representation Learning, Regression |
| Learning Methods | Self-Supervised Learning, End-to-End Learning, Representation Learning, Multi-Task Learning |
| Performance Highlights | validity: 74.0%, novelty: 100%, uniqueness: 100%, NRMSE_in-distribution_examples: 3.3%–3.6% (Fig.3a), 4.6% (best in Fig.3b) with variations up to 8.5%, comparison_best_training_match: 5.0% (closest existing design in training for Fig.3a), 7.7% (best match in training for Fig.3b), NRMSE_multi_target_examples: 4.7%–10.4% (for combined stress-strain + targeted Poisson’s ratios ν32 = -3.0, 0.0, 1.0), training_best_match_NRMSE: 11.2% (closest in training for the multi-target example), NRMSE_unseen_extreme_example: 2.2%–3.5% (for an unseen stress-strain + ν32 = -3.0 target that lies outside training distribution), training_best_match_NRMSE_for_unseen: 22.1% (best training candidate for that unseen target), NRMSE_unseen_cases: 7.2% and 7.0% (two unseen highly-nonlinear targets in Fig.4), training_best_match_NRMSE: 22.1% and 24.1% (best matches from training dataset for those targets), experimental_vs_FE_agreement: Qualitative/good agreement reported (no single aggregate numeric metric provided); representative stress-strain curves match key features (plateau, buckling-induced softening, contact-induced hardening) |
| Application Domains | Mechanical metamaterials / architected materials, Metamaterial inverse design, Materials discovery, Additive manufacturing / 3D printing (fabrication and experimental validation), Soft robotics (application example), Energy-absorbing components (application example), Protective gear / biomedical implants (multi-target design use-cases) |
302. Generative AI enables medical image segmentation in ultra low-data regimes, Nature Communications (July 14, 2025)
| Category | Items |
|---|---|
| Datasets | ISIC2018 (ISIC), PH2, DermIS, DermQuest, JSRT, NLM-MC, NLM-SZ, COVID-QU-Ex, FPD, FetReg, KVASIR, CVC-ClinicDB, BUID, FUSeg, ICFluid, ETAB, MSD Task04 (Hippocampus), MSD Task03 (Liver) |
| Models | U-Net, Generative Adversarial Network, Conditional GAN, Variational Autoencoder, Diffusion Model, Denoising Diffusion Probabilistic Model, Swin Transformer, Neural Architecture Search |
| Tasks | Semantic Segmentation, Semantic Segmentation, Semantic Segmentation |
| Learning Methods | Adversarial Training, End-to-End Learning, Supervised Learning, Semi-Supervised Learning, Neural Architecture Search, Gradient Descent |
| Performance Highlights | Jaccard_DermIS_GenSeg-UNet: 0.65, Jaccard_DermIS_UNet_baseline: 0.41, Jaccard_PH2_GenSeg-UNet: 0.77, Jaccard_PH2_UNet_baseline: 0.56, Dice_NLM-MC_GenSeg-UNet: 0.86, Dice_NLM-MC_UNet_baseline: 0.77, Dice_NLM-SZ_GenSeg-UNet: 0.93, Dice_NLM-SZ_UNet_baseline: 0.82, Jaccard_ISIC_GenSeg-SwinUnet: 0.62, Jaccard_ISIC_SwinUnet_baseline: 0.55, Jaccard_PH2_GenSeg-SwinUnet: 0.65, Jaccard_PH2_SwinUnet_baseline: 0.56, Jaccard_DermIS_GenSeg-SwinUnet: 0.62, Jaccard_DermIS_SwinUnet_baseline: 0.38, Absolute_gain_GenSeg-UNet_placental_vessels: 15%, Absolute_gain_GenSeg-UNet_skin_lesions: 9.6%, Absolute_gain_GenSeg-UNet_polyps: 11%, Absolute_gain_GenSeg-UNet_intraretinal_cystoid: 6.9%, Absolute_gain_GenSeg-UNet_foot_ulcers: 19%, Absolute_gain_GenSeg-UNet_breast_cancer: 12.6%, Placental_vessel_Dice_GenSeg-DeepLab_with_50_examples: 0.51 (GenSeg-DeepLab achieved similar to DeepLab with 500 examples per paper text), Foot_ulcer_Dice_GenSeg-UNet_with_50_examples: 0.6 (approx as reported contextual example; GenSeg-UNet required 50 vs UNet 600 to reach ~0.6), Lung_Dice_GenSeg-UNet_with_9_examples: 0.97 (paper states achieving Dice 0.97 required 175 examples for UNet, whereas GenSeg-UNet needed just 9 examples), Comparison_BBDM_End2End_vs_Pix2Pix_End2End: BBDM (End2End) achieved the highest performance across datasets; Pix2Pix (End2End) and Soft-Intro VAE (End2End) comparable but slightly lower, Computational_cost_BBDM_vs_Pix2Pix: BBDM incurs significantly higher computational cost and larger model size compared to Pix2Pix |
| Application Domains | Medical image analysis / medical imaging, Dermatology (skin lesion segmentation from dermoscopy), Pulmonology / Radiology (lung segmentation from chest X-ray), Obstetrics / Fetoscopy (placental vessel segmentation), Gastroenterology (polyp segmentation from colonoscopy), Wound care / Dermatology (foot ulcer segmentation), Ophthalmology (intraretinal cystoid fluid segmentation from OCT), Cardiology (left ventricle and myocardial wall segmentation from echocardiography), Breast imaging / Oncology (breast cancer segmentation from ultrasound), Neuroimaging and abdominal imaging (3D hippocampus and liver segmentation from MR/CT) |
301. La-Proteina: Atomistic Protein Generation via Partially Latent Flow Matching, Preprint (July 13, 2025)
| Category | Items |
|---|---|
| Datasets | Foldseek Clustered AFDB (filtered), Custom AFDB subset for long-length training, PDB (reference set samples), AFDB reference subsets for evaluation |
| Models | Variational Autoencoder, Transformer, Normalizing Flow, Diffusion Model, Encoder-Decoder, Autoencoder |
| Tasks | Synthetic Data Generation, Data Generation, Structured Prediction, Regression, Multi-class Classification, Clustering, Synthetic Data Generation |
| Learning Methods | Unsupervised Learning, Generative Learning, Pre-training, Representation Learning, Batch Learning |
| Performance Highlights | sequence_recovery_rate: 1.0 (perfect), average_all-atom_RMSD_reconstruction: ≈0.12 Å, co-designability_all-atom_%: 68.4, co-designability_alpha-carbon_%: 72.2, diversity_structure_clusters: 206, diversity_sequence_clusters: 216, diversity_str+seq_clusters: 301, novelty_TMScore_PDB: 0.75, novelty_TMScore_AFDB: 0.82, designability_MPNN-8%: 93.8, designability_MPNN-1%: 82.6, secondary_alpha_%: 72, secondary_beta_%: 5, co-designability_all-atom_%: 75.0, co-designability_alpha-carbon_%: 78.2, diversity_structure_clusters: 129, diversity_sequence_clusters: 199, diversity_str+seq_clusters: 247, novelty_TMScore_PDB: 0.82, novelty_TMScore_AFDB: 0.86, designability_MPNN-8%: 94.6, designability_MPNN-1%: 84.6, secondary_alpha_%: 73, secondary_beta_%: 6, La-Proteina (η_x, η_z)=(0.2,0.1) co-designability_all-atom_%: 60.6, La-Proteina (η_x, η_z)=(0.3,0.1) co-designability_all-atom_%: 53.8, overall_unconditional_generation_up_to_length: co-designable proteins up to 800 residues (La-Proteina remains viable where baselines collapse), co-designability_%: 21.2, diversity_all-atom_clusters: 51, diversity_seq_clusters: 105, diversity_str+seq_clusters: 91, KL=1e-3 co-designability_%: 65.2, KL=1e-4 co-designability_%: 83.8, KL=1e-5 co-designability_%: 82.4, best_co-designability_% (exp/quad scheduling, 0.1/0.1 noise): 68.4, other viable combinations co-designability_%: [60.6, 57.4, 59.2, 57.0, 54.0, 52.4, 50.6, 53.6, 55.4], tasks_solved_by_La-Proteina_out_of_26: 21-25 (depending on setup: all-atom vs tip-atom, indexed vs unindexed), tasks_solved_by_Protpardelle_out_of_26: 4, inference_time_batchsize1_seconds_length100: 2.94, inference_time_batchsize1_seconds_length200: 3.0, inference_time_batchsize1_seconds_length300: 3.67, inference_time_batchsize1_seconds_length400: 4.75, inference_time_batchsize1_seconds_length500: 6.33, inference_time_batchsize1_seconds_length600: 8.45, inference_time_batchsize1_seconds_length700: 10.63, inference_time_batchsize1_seconds_length800: 13.52, max_batch_inference_time_per_sample_length100: 0.34, max_batch_inference_time_per_sample_length800: 12.31 |
| Application Domains | Protein design / computational structural biology, De novo protein structure generation, Atomistic motif scaffolding (enzyme active site design, binder design implications), Biophysical / structural validation (rotamer modeling, MolProbity assessment) |
300. AlphaGenome: advancing regulatory variant effect prediction with a unified DNA sequence model, Preprint (July 11, 2025)
| Category | Items |
|---|---|
| Datasets | ENCODE, GTEx (via RECOUNT3), FANTOM5 (CAGE), 4D Nucleome (contact maps / Hi-C / Micro-C), PolyA_DB / Polyadenylation annotations, ClinVar, MFASS (Multiplexed Functional Assay of Splicing using Sort-seq), CAGI5 MPRA saturation mutagenesis challenge, Open Targets (GWAS credible sets), eQTL Catalog / SuSiE fine-mapped eQTLs, ChromBPNet benchmarks (caQTL/dsQTL/bQTL), ENCODE-rE2G (CRISPRi enhancer-gene validation), gnomAD common variants (chr22 subset) |
| Models | Transformer, U-Net, Convolutional Neural Network, Multi-Layer Perceptron, Multi-Head Attention, Self-Attention Network |
| Tasks | Regression, Binary Classification, Sequence Labeling, Structured Prediction, Link Prediction, Ranking, Feature Extraction / Representation Learning |
| Learning Methods | Supervised Learning, Pre-training, Knowledge Distillation, Ensemble Learning, Multi-Task Learning, Fine-Tuning, Representation Learning, Batch Learning, Gradient Descent |
| Performance Highlights | genome_track_evaluations_outperform_count: AlphaGenome outperformed external models on 22 out of 24 genome track evaluations, variant_effect_evaluations_outperform_count: AlphaGenome matched or outperformed external models on 24 out of 26 variant effect prediction evaluations, gene_expression_LFC_rel_improvement_vs_Borzoi: +17.4% (relative improvement in cell type-specific gene-level expression LFC Pearson r vs Borzoi), contact_maps_rel_improvement_vs_Orca_Pearson_r: +6.3% (Pearson r), +42.3% (cell type-specific differences), ProCapNet_rel_improvement_total_counts_Pearson_r: +15% (vs ProCapNet), ChromBPNet_rel_improvement_accessibility_Pearson_r: +8% ATAC, +19% DNase (total counts Pearson r), splice_benchmarks_SOTA: AlphaGenome achieves SOTA on 6 out of 7 splicing VEP benchmarks, ClinVar_deep_intronic_auPRC: 0.66 (AlphaGenome composite) vs 0.64 (Pangolin), ClinVar_splice_region_auPRC: 0.57 (AlphaGenome) vs 0.55 (Pangolin), ClinVar_missense_auPRC: 0.18 (AlphaGenome) vs 0.16 (DeltaSplice/Pangolin/DeltaSplice), MFASS_auPRC: 0.54 (AlphaGenome) vs 0.51 (Pangolin); SpliceAI/DeltaSplice = 0.49, Junctions_prediction_Pearson_r_examples: High correlations reported for junction counts across tissues (e.g., Pearson r ~0.75-0.76 in examples), contact_map_Pearson_r_vs_Orca: +6.3% Pearson r improvement; cell type differential prediction improvement +42.3% (compared to Orca), contact_map_examples_Pearson_r_values: Example intervals: AlphaGenome Pearson r ~0.79-0.86 vs ground truth maps (figure examples), zero_shot_causality_auROC_comparable_to_Borzoi: AlphaGenome zero-shot causality comparable to Borzoi (mean auROC ~0.68), supervised_RF_auROC: Random Forest using AlphaGenome multimodal features improved mean auROC from 0.68 (zero-shot) to 0.75, surpassing Borzoi supervised performance (mean auROC 0.71), zero_shot_cell_type_matched_DNase_Pearson_r: 0.57 (AlphaGenome cell type-matched DNase predictions; comparable to ChromBPNet and Borzoi Ensemble), LASSO_multi-celltype_DNase_Pearson_r: 0.63 (AlphaGenome with LASSO aggregation over all cell types), LASSO_multimodal_Pearson_r: 0.65 (AlphaGenome integrating multiple modalities across cell types; SOTA on CAGI5 reported), ENCODE-rE2G_zero_shot_auPRC: AlphaGenome outperformed Borzoi in identifying validated enhancer-gene links, particularly beyond 10 kb distance; zero-shot within 1% auPRC of ENCODE-rE2G-extended trained model, supervised_integration_auPRC_improvement: Including AlphaGenome features into ENCODE-rE2G-extended model increased auPRC to new SOTA across distance bins (Fig.4j), APA_Spearman_r: 0.894 (AlphaGenome) vs 0.790 (Borzoi) for APA prediction; reported as SOTA, paQTL_auPRC_within_10kb: 0.629 (AlphaGenome) vs 0.621 (Borzoi), paQTL_auPRC_proximal_50bp: 0.762 (AlphaGenome) vs 0.727 (Borzoi), caQTL_African_coefficient_Pearson_r: 0.74 (AlphaGenome predicted vs observed effect sizes for causal caQTLs; DNase GM12878 track example), SPI1_bQTL_coefficient_Pearson_r: 0.55 (AlphaGenome predicted vs observed SPI1 bQTLs), caQTL_causality_AP_mean: AlphaGenome achieved higher Average Precision vs Borzoi and ChromBPNet across multiple ancestries and datasets (specific AP values shown in Supplementary/Extended Data; e.g., AP = 0.50-0.63 depending on dataset), inference_speed: <1 second per variant on NVIDIA H100 (single student model), enabling fast large-scale scoring, overall_variant_benchmarks_outperform_count: Matched or outperformed external SOTA on 24/26 variant effect prediction benchmarks (Fig.1e) |
| Application Domains | Regulatory genomics, Variant effect prediction / clinical variant interpretation, Splicing biology and splicing variant interpretation, Gene expression regulation and eQTL interpretation, Alternative polyadenylation (APA) and paQTLs, Chromatin accessibility and TF binding QTL analysis, 3D genome architecture (contact map prediction), Enhancer–gene linking and functional genomics perturbation interpretation, Massively parallel reporter assay (MPRA) analysis, GWAS interpretation and prioritization |
298. Artificial Intelligence Paradigms for Next-Generation Metal–Organic Framework Research, Journal of the American Chemical Society (July 09, 2025)
| Category | Items |
|---|---|
| Datasets | Cambridge Structural Database (CSD) - MOF subset, Trillions of hypothetical MOF structures (unnamed hypothetical databases), CoRE MOF (Computation-ready, experimental MOF database), QMOF, MOFkey, DigiMOF, MOSAEC-DB, ARC-MOF, MOFX-DB, OpenDAC2023 dataset, ImageNet, MOFSimplify dataset (stability data) |
| Models | Transformer, Multi-Layer Perceptron, Recurrent Neural Network, Graph Neural Network, Graph Convolutional Network, Message Passing Neural Network, Convolutional Neural Network, Generative Adversarial Network, Variational Autoencoder, Gaussian Process, Attention Mechanism |
| Tasks | Regression, Node Classification, Graph Generation, Text Summarization, Text Classification, Information Retrieval, Hyperparameter Optimization, Feature Extraction, Representation Learning |
| Learning Methods | Supervised Learning, Self-Supervised Learning, Transfer Learning, Fine-Tuning, Reinforcement Learning, Unsupervised Learning, Representation Learning, Embedding Learning, Incremental Learning, Few-Shot Learning |
| Performance Highlights | accuracy_description: high-fidelity partial charge assignment, runtime: orders of magnitude shorter runtime compared to DFT, application: virtual screening for toluene vapor adsorption, hyperparameter_optimization_method: Bayesian optimization (mentioned), qualitative: high-accuracy gas adsorption predictions reported in transformer-based approaches (refs), examples: MOFormer (self-supervised), MOF-Transformer, multi-modal pre-training transformer for universal transfer learning, application: prediction of adsorption properties via 3D voxelized potential-energy-surfaces and nanoporous material recognition, qualitative: used for inverse design (property-to-structure) enabling targeted materials design, contextual_note: no numeric performance metrics reported in text, qualitative: facilitates inverse design; no explicit quantitative metrics reported in the perspective, qualitative: Gaussian regression or kernel methods presented as approaches to map atomic positions to potential energy surfaces within ML potentials context, application: MOF-GRU predicted gas separation performance (ref.72), quantitative_metrics: not provided in text, application: interpretable graph transformer network for predicting adsorption isotherms of MOFs (Ref.77), quantitative_metrics: not stated in this perspective, accuracy_description: near-quantum mechanical accuracy (qualitative), scaling: enable simulation of experimental-size MOF membranes (up to 28.2 × 28.2 nm^2) with high quality, application_example: predicted adsorption isotherm at 77 K via Grand Canonical Monte Carlo in excellent agreement with experimental data (case study) |
| Application Domains | metal-organic frameworks (MOF) materials discovery, gas storage and separation (CO2 capture, methane, hydrogen, toluene vapor adsorption), direct air capture (DAC) and sorbent discovery, catalysis, drug delivery / biomedical MOFs, water purification, environmental remediation, renewable energy and energy storage (batteries), materials synthesis optimization and autonomous laboratories, computational materials databases and information retrieval |
297. Accelerated data-driven materials science with the Materials Project, Nature Materials (July 03, 2025)
| Category | Items |
|---|---|
| Datasets | Materials Project (MP) main database, Matbench suite, Open Catalyst 2020 (OC20), Electronic charge density database (representation-independent), X-ray absorption spectra (XANES / EXAFS) dataset in MP, Ab initio non-crystalline structure database |
| Models | Graph Neural Network, Message Passing Neural Network, Random Forest, Attention Mechanism, Graph Neural Network |
| Tasks | Regression, Classification, Density Estimation, Graph Generation |
| Learning Methods | Supervised Learning, Semi-Supervised Learning, Unsupervised Learning, Active Learning, Pre-training, Contrastive Learning |
| Performance Highlights | None |
| Application Domains | materials science (inorganic crystals), energy storage / batteries (cathodes, solid electrolytes), catalysis / surface chemistry, optoelectronics / transparent conductors, phosphors / lighting materials, thermoelectrics, piezoelectrics, magnetocalorics, carbon capture materials, quantum materials / 2D systems / topological materials, non-crystalline / amorphous materials |
296. Natural-Language-Interfaced Robotic Synthesis for AI-Copilot-Assisted Exploration of Inorganic Materials, Journal of the American Chemical Society (July 02, 2025)
| Category | Items |
|---|---|
| Datasets | benchmark dataset of inorganic synthesis examples, solution-based inorganic materials synthesis procedures dataset, material characterization / crystallographic data |
| Models | GPT, Transformer |
| Tasks | Sequence-to-Sequence, Language Modeling, Information Retrieval, Decision Making, Planning, Experimental Design, Optimization |
| Learning Methods | Pre-training, Prompt Learning, In-Context Learning |
| Performance Highlights | syntax_validation_success_rate: ≈97%, semantic_fidelity_success_rate: ≈86%, iterations_for_syntax_validation: within three iterations, predefined_operations_coverage: 80% of procedures in inorganic synthesis dataset (ref. 54), materials_discovery_outcomes: synthesized 13 compounds across four classes; discovered new Mn−W clusters (Mn4W18, Mn4W8, Mn8W26, Mn57W42, new morphology of Mn72W48), human-AI_interaction_rounds_for_Mn-W_exploration: 65 rounds, new_structures_discovered: 4 structurally related new Mn−W clusters and 1 new morphology (Mn4W18, Mn4W8, Mn8W26, Mn57W42, new morphology of Mn72W48) |
| Application Domains | inorganic materials synthesis, chemical synthesis automation, robotic laboratory automation, materials discovery (polyoxometalates, metal−organic frameworks, nanoparticles, coordination complexes), experimental design and planning, human–AI collaborative research |
295. Self-Evolving Discovery of Carrier Biomaterials with Ultra-Low Nonspecific Protein Adsorption for Single Cell Analysis, Advanced Materials (July 02, 2025)
| Category | Items |
|---|---|
| Datasets | Experimental protein adsorption dataset (polyacrylamide copolymer formulations), Combinatorial formulation search space (design space), RDKit monomer descriptor dataset (computed descriptors for monomers), Detection sensitivity datasets (ELISA on plates and beads), Algorithm training / testing runs (SEBO training & tests) |
| Models | Random Forest |
| Tasks | Optimization, Experimental Design, Regression, Feature Selection, Hyperparameter Optimization |
| Learning Methods | Supervised Learning, Evolutionary Learning, Ensemble Learning, Hyperparameter Optimization |
| Performance Highlights | training_fitness_convergence: 0.1, training_datapoints_used_for_parameter_evolution: 10 |
| Application Domains | single-cell analysis, biomaterials discovery, protein analysis (ELISA detection sensitivity), microfluidics (bead carriers and plate carriers), automated experiments / autonomous laboratory workflows, materials science (copolymer discovery) |
294. El Agente: An autonomous agent for quantum chemistry, Matter (July 02, 2025)
| Category | Items |
|---|---|
| Datasets | SST |
| Models | None |
| Tasks | Sentiment Analysis |
| Learning Methods | None |
| Performance Highlights | None |
| Application Domains | Natural Language Processing, Sentiment Analysis |
293. Zero shot molecular generation via similarity kernels, Nature Communications (July 01, 2025)
| Category | Items |
|---|---|
| Datasets | QM9, SPICE, GEOM-Drug, SiMGen reference subsets (QM9-derived), Generated structures / supporting dataset |
| Models | Denoising Diffusion Probabilistic Model, Graph Neural Network, Message Passing Neural Network, Radial Basis Function Network, Multi-Layer Perceptron |
| Tasks | Data Generation, Graph Generation, Conditional generation, Synthetic Data Generation |
| Learning Methods | Self-Supervised Learning, Pre-training, Transfer Learning, Evolutionary Learning |
| Performance Highlights | training_data: trained on 80% of QM9 for 300 epochs, model_hyperparams: cutoff=10 Å, 16 radial basis functions, 2 interaction layers, 64 channels, message equivariance L=1, correlation ν=3, reference_efficiency: SiMGen can match trained-model performance using just 256 reference molecules (see SiMGen / 256 row in Table 1), MACE_pretraining_data: SPICE dataset (1 million molecules), descriptor_cutoff: 5.0 Å, MACE_descriptor_channels: first layer invariant scalar node features used, penicillin_guidance_fold_change_β-lactam: ≈8× increase, penicillin_guidance_fold_change_thiolane: ≈3× increase, hydrogenation_validity_before: ≈99% valid atoms before hydrogen addition across sizes, generation_scaling: SiMGen generated molecules with 5-50 heavy atoms; validity after hydrogenation decreases slowly with size; with open priors valid molecules ≈0.6 for 50 heavy atoms, energy_based_time_encoding: time is positionally encoded and combined via MLP into node features for the energy-based MACE model |
| Application Domains | de novo molecular generation, drug discovery (fragment-based design, linker design, binder design), materials design / crystal structure generation (discussion and motivation), molecular force-field modelling and force prediction, interactive molecular design tools (ZnDraw web tool) |
292. Enabling large language models for real-world materials discovery, Nature Machine Intelligence (July 2025)
| Category | Items |
|---|---|
| Datasets | Battery Device QA, MaScQA, MatSciNLP, OpticalTable / OpticalTable-SQA, SustainableConcrete, NanoMine, ChatExtract, Structured Information Extraction, MatText, MaCBench, LitQA, RedPajama (training corpus reference), Materials Project, DPA-2 (large atomic model dataset/model), Open Reaction Database, Polymer nanocomposite data (ACS Macro Lett.) |
| Models | BERT, GPT, Transformer, Vision Transformer, Graph Neural Network, Diffusion Model, Denoising Diffusion Probabilistic Model, Multi-Layer Perceptron |
| Tasks | Named Entity Recognition, Question Answering, Text Classification, Sequence-to-Sequence, Regression, Image Generation, Graph Generation, Planning, Decision Making, Information Retrieval, Data Generation |
| Learning Methods | Pre-training, Fine-Tuning, In-Context Learning, Prompt Learning, Transfer Learning, Multi-Agent Learning, Reinforcement Learning, Representation Learning |
| Performance Highlights | questions_examined: 650 |
| Application Domains | Materials science (general), Chemistry (adjacent domain and source of methods), Batteries / energy materials, Optical materials, Concrete / civil materials, Nanomaterials, Computational materials science (in silico evaluation), Automated experimentation / laboratory robotics, Sustainable materials and manufacturing, Bio-inspired materials / biological materials (briefly referenced) |
291. A framework for evaluating the chemical knowledge and reasoning abilities of large language models against the expertise of chemists, Nature Chemistry (July 2025)
| Category | Items |
|---|---|
| Datasets | ChemBench (full corpus), ChemBench-Mini, Chemical preference dataset (Choung et al. 2023) — sampled subset, MoleculeNet (referenced), Therapeutics Data Commons (referenced), MatBench (referenced), PubChem and Gestis (referenced as specialized databases) |
| Models | Transformer, GPT |
| Tasks | Question Answering, Multi-class Classification, Text Generation, Regression, Classification, Ranking, Uncertainty / Confidence estimation |
| Learning Methods | Self-Supervised Learning, Fine-Tuning, Few-Shot Learning, Prompt Learning, Pre-training |
| Performance Highlights | fraction_correct: 0.71, fraction_correct: 0.22, alignment_with_experts: often indistinguishable from random guessing, example_confidence_reported: GPT-4: reported 1.0 for one correct answer and 4.0 for six incorrect answers (on 1–5 scale), note: PaperQA2 (agentic system) included in evaluation; relative performance shown in Fig. 3 (no single-number reported in main text) |
| Application Domains | chemistry, analytical chemistry, organic chemistry, inorganic chemistry, physical chemistry, technical chemistry, materials science, medicinal chemistry / drug discovery, chemical safety / toxicity assessment, chemical education / assessment |
290. A generalized platform for artificial intelligence-powered autonomous enzyme engineering, Nature Communications (July 01, 2025)
| Category | Items |
|---|---|
| Datasets | AtHMT variant screening dataset (this study, Supplementary Data 3), YmPhytase variant screening dataset (this study, Supplementary Data 4), Initial variant libraries (this study), ESM-2 pretraining corpus (referenced), Multiple sequence alignments / homologous sequences used by EVmutation |
| Models | Transformer, Markov Random Field, GPT, Supervised Learning |
| Tasks | Regression, Data Generation, Optimization, Experimental Design, Sequence-based prediction / zero-shot fitness prediction (mapped to Regression/Zero-Shot Learning) |
| Learning Methods | Unsupervised Learning, Zero-Shot Learning, Supervised Learning, Self-Supervised Learning, Prompt Learning, Representation Learning |
| Performance Highlights | best_single_mutant_fold_change_AtHMT: 2.1, best_single_mutant_fold_change_YmPhytase: 2.6, initial_library_percent_variants_above_wt_AtHMT: 59.6%, initial_library_percent_variants_above_wt_YmPhytase: 55%, initial_library_percent_significantly_better_AtHMT: 50% (of above-wt?), initial_library_percent_significantly_better_YmPhytase: 23% (two-tailed Student’s t-test p<0.05), relative_performance_vs_ESM2: EVmutation performed better overall than ESM-2 on initial predictions (qualitative statement), overlap_between_models_predictions: substantial overlap (qualitative), screened_variants_per_round_constructed: top 96 predicted mutants constructed (90 used for screening), third_round_model_predicted_triple_mutants_better_than_V140T: 74/90 (82%), human_intuited_S99T/V140T_triple_mutants_better_than_V140T: 4/36 (11%), general_correlation_with_experimental_results: relatively weak / little overall consistency (qualitative) |
| Application Domains | Protein engineering, Synthetic biology, Biocatalysis, Metabolic engineering, Natural product discovery, Biotechnology, Medicine (enzyme applications), Renewable energy and sustainable chemistry (industrial enzyme applications) |
289. Machine-learning design of ductile FeNiCoAlTa alloys with high strength, Nature (July 2025)
| Category | Items |
|---|---|
| Datasets | Training dataset of FCC HEAs (AlCoCrFeNiTa system), Experimental synthesis / validation dataset (iterative active-learning candidates), Atom probe tomography / SAXS measurement datasets (characterization data) |
| Models | not specified (surrogate machine learning model) |
| Tasks | Regression, Optimization, Experimental Design, Ranking |
| Learning Methods | Active Learning, Supervised Learning |
| Performance Highlights | ML_prediction_accuracy: not reported (numerical ML metrics such as RMSE/R² not provided in main text), experimental_outcome_yield_strength: 1.75 GPa ± 0.05 GPa (representative HEA05 after 750 °C for 1 h aging), experimental_outcome_ultimate_tensile_strength: 2.403 GPa ± 0.046 GPa, experimental_outcome_uniform_elongation: 25% ± 1.5%, work_hardening_rate: > 2 GPa (stable across wide strain range), true_stress_peak: ≈ 3 GPa (true stress), performance_range_sigma_y_vs_eu: σy range 1.5–1.95 GPa with εu range 31%–15% reported across processing variants, σUTS_minus_σy: 650 MPa (work hardening capability example), yield_ratio_σy/σUTS: 0.73 (example), σUTS_times_εf: ≈ 60 GPa% (representative) |
| Application Domains | Materials science, Metallurgy, High-entropy alloy design, Mechanical engineering (structural materials) |
288. Tracking 35 years of progress in metallic materials for extreme environments via text mining, Scientific Reports (July 01, 2025)
| Category | Items |
|---|---|
| Datasets | Web of Science corpus (titles and abstracts) collected for “metallic materials in extreme environments”, Pre-trained word-embedding model corpus from Pei et al. (used as benchmark / transfer-learning starting point) |
| Models | Latent Dirichlet Allocation, BERT, GPT |
| Tasks | Representation Learning, Information Retrieval, Recommendation, Clustering, Feature Extraction, Named Entity Recognition |
| Learning Methods | Self-Supervised Learning, Transfer Learning, Fine-Tuning, Unsupervised Learning, Pre-training, Representation Learning |
| Performance Highlights | num_topics: 7, no_above: 0.5, passes: 15, iterations: 400, vector_dimension: 200, context_window: 8, epochs: 30, period_counts: 1989–2003:157,451;2004–2009:128,026;2010–2014:146,382;2015–2018:171,475;2019–2021:186,900;2022–2023:141,124, example_rankings: Ti-Nb-Zr ranked 222 (2004–2009); Ti-13Nb-13Zr ranked 381 (2004–2009), evaluation_metric_used: cosine similarity / cosine distance (reported; values shown in figures but not explicitly numerically reported in main text), alignment_method: Orthogonal Procrustes (SVD: Y^T X = U Σ V^T; O = U V^T); vectors normalized after transform, goal: make embeddings from different time periods comparable; cosine distances used for term association |
| Application Domains | Metallic materials for extreme environments, Materials science (high-entropy alloys / multi-principal-element alloys), Aerospace engineering, Biomedical engineering, Renewable energy (battery research, wind turbines, solar panels), Nuclear / irradiation environments, Gas pipelines / hydrogen transport |
286. Large language models to accelerate organic chemistry synthesis, Nature Machine Intelligence (July 2025)
| Category | Items |
|---|---|
| Datasets | USPTO-50k, Open Reaction Database (ORD), Suzuki–Miyaura (HTE) dataset, Imidazole C–H arylation (HTE) dataset, Buchwald–Hartwig (ELN) dataset, Regioselectivity dataset (Li et al.), Enantioselectivity dataset (Zahrt et al.), Pd-catalysed carbonylation literature dataset, Curated Q&A training dataset (Chemma) |
| Models | Transformer, GPT, Seq2Seq, Multi-Layer Perceptron, Feedforward Neural Network, Random Forest, Graph Neural Network, Gaussian Process |
| Tasks | Sequence-to-Sequence, Retrosynthesis, Forward prediction, Condition generation, Regression, Recommendation, Optimization, Active Learning, Selectivity (as regression), Image/other tasks not applicable |
| Learning Methods | Supervised Learning, Fine-Tuning, Pre-training, Reinforcement Learning, In-Context Learning, Zero-Shot Learning, Active Learning, Transfer Learning |
| Performance Highlights | top-1_accuracy: 72.2%, Suzuki–Miyaura (example): R2 = 0.86, RMSE = 5.20%, Suzuki–Miyaura (other splits): R2 = 0.85, RMSE = 5.40%, Buchwald–Hartwig (ELN): R2 = 0.79, RMSE = 6.56%, Imidazole C–H arylation: R2 = 0.74, RMSE = 6.59%, Another reported value for imidazole C–H arylation (figure): R2 = 0.83, RMSE = 6.02%, Buchwald–Hartwig (other figure): R2 = 0.81, RMSE = 5.51%, Regioselectivity: R2 = 0.93, RMSE = 0.74 kcal mol−1, site_accuracy = 78.74%, Enantioselectivity (chiral phosphoric acid catalysed thiol addition): R2 = 0.89, RMSE = 0.25 kcal mol−1 (Chemma) ; comparison: Li et al. reported R2 = 0.915, RMSE = 0.197 kcal mol−1, Ligand_recommendation_median_performance: For 15 of the 16 base–solvent combinations, the recommended ligand performs best in terms of median reaction yields (paper summary statistic), Chemma-enhanced_RF (5% real + generated data) Suzuki–Miyaura: R2 = 0.53, Chemma-enhanced_RF (5% real + generated data) Buchwald–Hartwig: R2 = 0.72, RF with 90% real data (reference): approx R2 = 0.6 (Suzuki) and 0.8 (Buchwald) as baseline reported in paper, Optimization_speed_Suzuki–Miyaura: Chemma-BO achieves 98.5% yield within first 15 experiments (3 batches) vs BO and GPT-4 requiring ~50 experiments, Optimization_speed_Buchwald–Hartwig: Chemma-BO achieves 98.7% within first 10 experiments; within first 25 experiments reaches 99.8% yield while BO requires at least 50 experiments |
| Application Domains | Organic chemistry synthesis, Drug discovery (medicinal chemistry / synthesis planning), Materials and energy (catalyst design and synthesis), Automated / autonomous experimentation (robotic chemistry and HTE integration) |
285. UMA: A Family of Universal Models for Atoms, Preprint (June 30, 2025)
| Category | Items |
|---|---|
| Datasets | OMat24, OMol25 (OMol-preview used in main training), OC20++ (OC20 All + MD + Rattled + clean surface + OC20-Multi-Adsorbate mAds), OMC25, ODAC25 (subset overlapping ODAC23), Combined UMA training corpus, MPTrj (fine-tuning), sAlex (fine-tuning) |
| Models | Graph Neural Network, Message Passing Neural Network, Multi-Layer Perceptron, Transformer |
| Tasks | Regression, Classification, Ranking, Optimization, Image/graph matching (mapped to Graph Matching) |
| Learning Methods | Supervised Learning, Multi-Task Learning, Pre-training, Fine-Tuning, Transfer Learning, Representation Learning |
| Performance Highlights | Materials Energy/Atom (meV): 20.0, Materials Forces (meV/Å): 60.8, Materials Stress (meV/Å^3): 4.4, Matbench F1: 0.916, AdsorbML Success Rate: 68.35%, OMol25 Ligand-strain MAE (meV): 4.39, CSP Lattice Energy MAE (kJ/mol): 2.695, Inference (1000 atoms) steps/sec: 16, Materials Energy/Atom (meV): 18.1, Materials Forces (meV/Å): 51.4, Materials Stress (meV/Å^3): 4.3, Matbench F1: 0.93, AdsorbML Success Rate: 71.12%, OMol25 Ligand-strain MAE (meV): 2.45, CSP Lattice Energy MAE (kJ/mol): 2.664, ODAC Test Ads. Energy (meV): 290.2, Inference (1000 atoms) steps/sec: 3, Materials Energy/Atom (meV): 17.6, Materials Forces (meV/Å): 45.5, Materials Stress (meV/Å^3): 3.8, Matbench F1: 0.928, AdsorbML Success Rate: 74.41% (25% improvement in successful adsorption energy calculations reported for catalysis vs previous SOTA), OMol25 Ligand-strain MAE (meV): 3.37, CSP Lattice Energy MAE (kJ/mol): 2.488, ODAC Test Ads. Energy (meV): 291.1, Inference (1000 atoms) steps/sec: 1.6, MoLE vs Dense compute-optimal gain (∆): ≈2.5× fewer active parameters for MoLE to achieve equivalent loss (reported for UMA-M), Validation loss behavior: MoLE models achieve lower validation loss at fixed FLOPs in experiments (Figures 3 & 4 discussed), AdsorbML previous SOTA success rate (EquiformerV2): ≈60.80% (literature baseline), UMA-L AdsorbML success rate: 74.41%, Example baseline OC20 Ads. Energy Force MAEs (GemNet / eqv2 reported values): GemNet-OC20: Ads. Energy 163.5 meV, Forces 16.3 meV/Å (literature column), UMA improvements: UMA models reduce OC20 adsorption energy errors by ~80% in some evaluations (paper statement) |
| Application Domains | computational chemistry, materials science, catalysis, drug discovery / structure-based drug design, energy storage (battery materials), semiconductor materials, molecular crystals / crystal structure prediction, metal-organic frameworks (MOFs) and direct-air capture applications, molecular dynamics simulations |
284. Rethinking chemical research in the age of large language models, Nature Computational Science (June 24, 2025)
| Category | Items |
|---|---|
| Datasets | MoleculeNet, Tox21, ChemBench, LAION-5B, QM9, Reaxys, SciFinder, Chatbot Arena (human preference data) |
| Models | Transformer, GPT, BERT, Graph Neural Network, Decision Tree, Random Forest, Autoencoder, Variational Autoencoder, Feedforward Neural Network, Decision Transformer, CLIP (implied), ChemLLM (domain-specific LLM) |
| Tasks | Planning, Optimization, Question Answering, Sequence-to-Sequence, Classification, Regression, Language Modeling, Text Generation |
| Learning Methods | Pre-training, Fine-Tuning, Reinforcement Learning, Transfer Learning, Prompt Learning, Knowledge Distillation, Self-Supervised Learning, Model-Based Reinforcement Learning (implied) |
| Performance Highlights | accuracy_Tox21: >80%, accuracy_translation: <10%, accuracy_yield_general: 70-80%, accuracy_yield_custom_methods: >95%, accuracy_reaction_product_general: ~20%, accuracy_reaction_product_bespoke: >90% |
| Application Domains | Chemistry, Chemical engineering, Materials science, Medicinal chemistry / drug discovery, Analytical chemistry (spectroscopy, NMR, mass spectrometry, IR), Automated experimentation / cloud labs / robotic laboratories, Knowledge management and ontology / knowledge graphs in scientific domains |
283. Agent-based multimodal information extraction for nanomaterials, npj Computational Materials (June 23, 2025)
| Category | Items |
|---|---|
| Datasets | DiZyme nanomaterials subset (test), DiZyme nanomaterials larger set (Jaccard evaluation), DiZyme nanozyme dataset, NER annotated corpus (training), NER annotated corpus (test), YOLO figure detection dataset |
| Models | GPT, Transformer, YOLO, BERT |
| Tasks | Named Entity Recognition, Object Detection, Structured Prediction, Sequence Labeling, Information Retrieval |
| Learning Methods | Fine-Tuning, Pre-training, Zero-Shot Learning, Gradient Descent, End-to-End Learning |
| Performance Highlights | Mw(coating)_precision_text_only: 0.62, Mw(coating)_precision_text+NER: 0.66, Mw(coating)_recall_text_only: 0.73, Mw(coating)_recall_text+NER: 0.86, nanoMINER_avg_precision: 0.89, nanoMINER_avg_recall: 0.72, nanoMINER_F1: 0.79, training_box_loss_end: 0.2, training_classification_loss_end: 0.17, trained_on_images: 537, GPT-4.1_avg_precision: 0.71, GPT-4.1_avg_recall: 0.65, GPT-4.1_F1: 0.68, o3-mini_avg_precision: 0.68, o3-mini_avg_recall: 0.57, o3-mini_F1: 0.62, o4-mini_avg_precision: 0.78, o4-mini_avg_recall: 0.69, o4-mini_F1: 0.74, crystal_system_inference_accuracy: 0.86, Cmin_precision_text_only: 0.9, Cmin_precision_text+vision: 0.97, Cmax_precision_text_only: 0.91, Cmax_precision_text+vision: 0.98, Km_precision_overall: 0.97, Vmax_precision_overall: 0.96, pH_precision_overall: 0.89, Temperature_precision_overall: 0.68, Km_recall_range: 0.87-0.91, Vmax_recall_range: 0.79-0.83, concentration_recall_range: 0.38-0.54 |
| Application Domains | Materials science (nanomaterials), Nanozymes / bionanotechnology, Chemistry, Biomedical data extraction (mentioned as extensible application), Scientific literature mining / knowledge base construction |
282. All-atom Diffusion Transformers: Unified generative modelling of molecules and materials, International Conference on Machine Learning (June 18, 2025)
| Category | Items |
|---|---|
| Datasets | MP20, QM9, GEOM-DRUGS, QMOF |
| Models | Variational Autoencoder, Autoencoder, Transformer, Denoising Diffusion Probabilistic Model, Autoencoder |
| Tasks | Data Generation, Synthetic Data Generation, Representation Learning, Distribution Estimation |
| Learning Methods | Unsupervised Learning, Self-Supervised Learning, Representation Learning, Transfer Learning, Batch Learning |
| Performance Highlights | MP20_joint_transformer_match_rate (%): 88.6, MP20_joint_transformer_RMSD (Å): 0.0239, QM9_joint_transformer_match_rate (%): 97.00, QM9_joint_transformer_RMSD (Å): 0.0399, MP20_structure_validity (%): 99.74, MP20_compositional_validity (%): 92.14, MP20_overall_validity (%): 91.92, MP20_metastable_rate (%): 81.0, MP20_stable_rate (%): 15.4, MP20_M.S.U.N. (%): 28.2, MP20_S.U.N. (%): 5.3, QM9_validity (%): 97.43, QM9_uniqueness (%): 96.92, PoseBusters_atoms_connected (%): 99.70, PoseBusters_bond_angles (%): 99.85, PoseBusters_bond_lengths (%): 99.41, PoseBusters_ring_flat (%): 100.00, PoseBusters_double_bond_flat (%): 99.98, PoseBusters_internal_energy (%): 95.86, PoseBusters_no_steric_clash (%): 99.79, GEOM-DRUGS_validity (%): 95.3, GEOM-DRUGS_uniqueness (%): 100.0, PoseBusters_valid (%): 85.3, PoseBusters_atoms_connected (%): 93.0, PoseBusters_ring_flat (%): 95.4, DiT-S_params: 32M, DiT-B_params: 130M (150M reported in some configs), DiT-L_params: 450M, Correlation_training_loss_vs_params_Pearson_at_epoch2000: -1.00, Correlation_crystal_validity_vs_params_Pearson: 0.91, Correlation_molecule_validity_vs_params_Pearson: 0.94, QMOF_only_validity_rate (%): 15.7, Joint_QMOF_validity_rate (%): 10.2, time_to_sample_10k_on_V100: under 20 minutes (ADiT reported), baseline_time_examples: equivariant baselines up to 2.5 hours on same hardware, speedup: Order-of-magnitude faster than equivariant diffusion baselines for 10k samples on single V100 |
| Application Domains | Generative chemistry, Materials design / inorganic crystals, Small-molecule design (drug-like molecules), Metal-organic frameworks (MOFs), Foundation models for atomic-scale structure generation |
281. Data-Driven Design of Random Heteropolypeptides as Synthetic Polyclonal Antibodies, Journal of the American Chemical Society (June 18, 2025)
| Category | Items |
|---|---|
| Datasets | RHP library for IFN campaign (384 randomly sampled candidates, 6 optimization iterations), RHP library for TNF-α campaign (524 randomly sampled candidates, 6 optimization iterations), ELISA measurement dataset (Target and Control signals used to compute Target, Control, and composite Score), BLI validation dataset (biolayer interferometry KD measurements), Functional neutralization assay dataset (L929 cell cytotoxicity, IC50 measurements), LP-EM single-molecule imaging dataset and MD simulation outputs |
| Models | Linear Model, Gaussian Process, Multi-Model (surrogate + evolutionary) |
| Tasks | Regression, Optimization, Experimental Design, Ranking |
| Learning Methods | Supervised Learning, Evolutionary Learning, Model-Based Learning |
| Performance Highlights | R2_Target_test: 0.7, R2_Control_test: 0.75, R2_Score_test: 0.25, R2_Target_train: 0.96, R2_Control_train: 0.97, R2_Score_train: 0.69, R2_Target_test: 0.89, R2_Control_test: 0.9, R2_Score_test: 0.4, SpAb_T1_KD_TNF-alpha: 7.9 nM, SpAb_T1_KD_HSA: 3.3 μM, Selectivity_TNF-alpha_over_HSA: ≈418-fold, SpAb_T1_KD_after_affinity_purification: <1.6 nM, SpAb_T2_KD_TNF-alpha: 413 nM, SpAb_T2_KD_HSA: 2.1 μM, SpAb_I2_KD_IFN: 103 nM, Neutralization_IC50_TNF-alpha_alone: 0.36 pg/mL, Neutralization_IC50_TNF-alpha_plus_SpAb_T1: 70 pg/mL, Neutralization_IC50_TNF-alpha_plus_RHP_T2: 0.37 pg/mL, Neutralization_IC50_TNF-alpha_plus_purified_SpAb_T1: 272 pg/mL, Neutralization_IC50_TNF-alpha_plus_anti-TNF-alpha_mAb: 1591 pg/mL |
| Application Domains | biomedicine, therapeutics, diagnostics, polymer materials discovery, biomolecular recognition, drug development, experimental automation / self-driving labs |
279. Agents for self-driving laboratories applied to quantum computing, Preprint (June 05, 2025)
| Category | Items |
|---|---|
| Datasets | Translation benchmark: 80 instructions from 8 experiments, Visual inspection synthetic dataset (four experiment types), LeeQ built-in experiments indexed by k-agents (benchmark & real runs), Real hardware experiment logs — single-qubit calibration, two-qubit siZZle parameter search, GHZ tomography |
| Models | GPT, Transformer, Gaussian Mixture Model |
| Tasks | Text Generation, Sequence-to-Sequence, Binary Classification, Image Classification, Clustering, Optimization, Experimental Design |
| Learning Methods | Few-Shot Learning, Zero-Shot Learning, Fine-Tuning, Prompt Learning, Pre-training, Representation Learning, Ensemble Learning |
| Performance Highlights | translation_accuracy: 97%, translation_accuracy_agent_based_GPT-4o(Agents): 99.17%, translation_accuracy_GPT-4o(LongContext): 97.92%, parameter_search_experiments_run: 100 experiments (3 hours), tested up to 20 frequencies, discovered_parameters: frequency 4726 MHz, amplitude 0.3049 (successful set reported), GHZ_state_fidelity: 83.83% |
| Application Domains | Quantum computing (superconducting quantum processors), Laboratory automation / self-driving laboratories, Scientific experiment execution and analysis (multimodal: text + images + code) |
278. SciAgents: Automating Scientific Discovery Through Bioinspired Multi-Agent Intelligent Graph Reasoning, Advanced Materials (June 05, 2025)
| Category | Items |
|---|---|
| Datasets | Ontological knowledge graph (global graph from prior work), Semantic Scholar search results (publication abstracts returned per query) |
| Models | GPT, Transformer |
| Tasks | Language Modeling, Text Generation, Question Answering, Text Summarization, Information Retrieval, Novelty Detection, Graph Generation, Data Generation |
| Learning Methods | In-Context Learning, Multi-Agent Learning, Prompt Learning, Pre-training, Fine-Tuning, Adversarial Training |
| Performance Highlights | document_length: 8100 words (example generated document), tensile_strength_prediction_for_proposed_material: up to 1.5 GPa (compared to traditional 0.5–1.0 GPa), energy_consumption_reduction: ~30% (projected for low-temperature processing), novelty_score_examples: Idea 1: 8, Idea 2: 8, Idea 3: 6, Idea 4: 7, Idea 5: 8 (Novelty / Feasibility pairs reported in Table 4), feasibility_score_examples: Idea 1: 7, Idea 2: 7, Idea 3: 8, Idea 4: 8, Idea 5: 7, MD_simulation_protocol_duration: 100–500 ns (protocol suggested by Critic agent for MD runs), MD_analysis_outputs: interaction energies, binding sites, cluster analysis of self-assembled structures (qualitative outputs suggested), general_claim: foundation LLMs (Transformer-based) provide strong generative capabilities but face accuracy and explainability challenges (qualitative) |
| Application Domains | bio-inspired materials, materials science, biomaterials, molecular modeling (molecular dynamics, DFT), synthetic biology, microfluidics, bioelectronics, generative materials informatics / scientific discovery automation, scientific literature mining / information retrieval |
277. A data-driven platform for automated characterization of polymer electrolytes, Matter (June 04, 2025)
| Category | Items |
|---|---|
| Datasets | PEO-salt polymer electrolyte dataset (this work) |
| Models | None |
| Tasks | Regression, Optimization, Feature Extraction, Feature Selection |
| Learning Methods | Supervised Learning, Active Learning |
| Performance Highlights | dataset_size: 70 unique formulations; 330 samples; ~2,000 ionic conductivity measurements, throughput: 67.5 samples per researcher hour (stated: “67.5 samples per researcher hour” / “over 60 samples per researcher hour”), HT_experiment_capacity: 90 electrolyte samples processed from start to finish in just under 5 days (includes 24 h drying downtime), measurement_precision_examples: in situ actuator thickness measurement inherent error < ±5 μm (quoted as ±5mm in text formatting), actuator position resolution 0.01 mm, force resolution 5 mN, temperature sensor 0.1 °C resolution |
| Application Domains | Battery materials / Electrolytes, Polymer electrolyte characterization, Materials science (experimental high-throughput data generation), Sodium-ion and Lithium-ion battery research, Data-driven materials discovery (enabling ML model training and optimization) |
276. IvoryOS: an interoperable web interface for orchestrating Python-based self-driving laboratories, Nature Communications (June 04, 2025)
| Category | Items |
|---|---|
| Datasets | None |
| Models | GPT, Convolutional Neural Network, Gaussian Process |
| Tasks | Optimization, Image Matching, Text Generation, Hyperparameter Optimization |
| Learning Methods | Prompt Learning, In-Context Learning, Model-Based Learning, Supervised Learning |
| Performance Highlights | None |
| Application Domains | Chemistry (self-driving laboratories, automated experimentation), Materials chemistry, Drug discovery, Formulation science, Laboratory automation and robotics, Analytical chemistry (HPLC, reaction monitoring), Automated synthesis and purification workflows |
275. An unsupervised machine learning based approach to identify efficient spin-orbit torque materials, npj Computational Materials (June 03, 2025)
| Category | Items |
|---|---|
| Datasets | APS and IEEE abstracts (1970-2020), Literature reports of spin Hall conductivities and measured SOT efficiencies |
| Models | Feedforward Neural Network, Transformer |
| Tasks | Representation Learning, Embedding Learning, Ranking, Regression, Clustering, Dimensionality Reduction, Information Retrieval, Feature Extraction |
| Learning Methods | Unsupervised Learning, Representation Learning, Embedding Learning |
| Performance Highlights | training_corpus_size_abstracts: Approximately 1,000,000 abstracts, embedding_dimension: 200, embedding_model_training_time: ~20 hours on Intel Xeon X5550, 24 GB RAM, new_candidates_identified: 97, high_SOT_candidates_predicted_xiNN>=1: 16, FeSi_experimental_xiSOT: 2 (experimental, Table 1), FeSi_xiNN_prediction: 1.82, example_Pt_experiment_vs_prediction: Pt ξ_SOT_exp=0.07, ξ_NN=0.07 (Table 1), example_Ta_experiment_vs_prediction: Ta ξ_SOT_exp=0.15, ξ_NN=0.42 (Table 1) |
| Application Domains | Materials science, Spintronics, Condensed matter physics, Device engineering (e.g., MRAM and nanomagnet switching), Scientific text mining / literature-based materials discovery |
274. Biomni: A General-Purpose Biomedical AI Agent, Preprint (June 02, 2025)
| Category | Items |
|---|---|
| Datasets | bioRxiv corpus (25 subject categories, 100 recent publications per category), Biomni-E1 environment (curated resources), LAB-Bench (subset used), Humanity’s Last Exam (HLE) (subset used), Open Targets genetics ground truth set (processed), GWAS causal gene detection dataset (Shringarpure et al.), CRISPR perturbation screen dataset (Schmidt et al.), scRNA-seq annotation datasets (various author-provided datasets), Microbiome benchmark datasets (5 datasets), Drug repurposing dataset (Huang et al.), Rare disease diagnosis dataset (MyGene2 curated by Alsentzer et al.), Patient gene prioritization dataset (Alsentzer et al.), Wearable sensor case-study data (CGM + body temperature), Wearable sleep data (case study), Multi-omics datasets merged with wearable data (case study), Human embryonic skeletal multi-omic atlas (To et al.) |
| Models | Transformer, GPT, Gradient Boosting Tree, Attention Mechanism |
| Tasks | Question Answering, Ranking, Optimization, Multi-class Classification, Recommendation, Clustering, Dimensionality Reduction, Experimental Design, Feature Extraction |
| Learning Methods | Zero-Shot Learning, In-Context Learning, Prompt Learning, Pre-training, Reinforcement Learning |
| Performance Highlights | DbQA_accuracy: 74.4%, SeqQA_accuracy: 81.9%, HLE_accuracy: 17.3%, relative_performance_gain_vs_base_LLM_avg_across_8_tasks: 402.3% (average relative gain), relative_gain_vs_coding_agent: 43.0%, relative_gain_vs_Biomni-ReAct: 20.4%, evaluation_metric: average post-perturbed effect (used to compare designed gene panels); specific numeric value not provided in paper for Biomni absolute score, cloning_benchmark_accuracy_vs_expert: Biomni matched human expert in accuracy and completeness across 10 realistic cloning tasks (scored by blinded expert using rubric); trainee-level human performed worse, wetlab_validation: Successful colonies on plates; Sanger sequencing of two picked colonies showed perfect alignment (successful insertion), GRN_findings: Recovered 566-589 regulons; identified known regulators (e.g., RUNX2) and novel regulators (AUTS2, ZFHX3, PBX1) with notable activity patterns, runtime: Full run completed in just over five hours (end-to-end for GRN analysis pipeline) |
| Application Domains | Biomedical research (general), Genetics, Genomics, Molecular biology, Single-cell biology / multi-omics, Microbiology / Microbiome analysis, Pharmacology / Drug repurposing, Clinical medicine / Rare disease diagnosis, Bioinformatics, Bioengineering, Biophysics, Pathology, Consumer health / wearable data analysis, Experimental wet-lab protocol design (molecular cloning) |
273. A multimodal conversational agent for DNA, RNA and protein tasks, Nature Machine Intelligence (June 2025)
| Category | Items |
|---|---|
| Datasets | Nucleotide Transformer benchmark (instructional version), Curated genomics instructions dataset (27 tasks) [created by this paper], APARENT2 dataset (polyadenylation), Saluki dataset (RNA degradation), ESM2 protein benchmark datasets (protein properties), AgroNT benchmark (plant genomes / enhancers), DeepSTARR dataset (enhancer activity in Drosophila), ChromTransfer dataset (regulatory element accessibility), BEND benchmark (subset used) |
| Models | Transformer, GPT, BERT, Convolutional Neural Network, Encoder-Decoder, Attention Mechanism, Cross-Attention, Self-Attention Network, Multi-Head Attention |
| Tasks | Binary Classification, Multi-label Classification, Regression, Sequence-to-Sequence, Language Modeling, Image Classification |
| Learning Methods | Self-Supervised Learning, Pre-training, Fine-Tuning, Multi-Task Learning, Supervised Learning, Transfer Learning, Backpropagation, End-to-End Learning, Representation Learning |
| Performance Highlights | MCC: 0.77 (ChatNT with English-aware projection, mean across 18 tasks), MCC_non_aware_projection: 0.71 (ChatNT with Perceiver resampler not conditioned on question), MCC_baseline_NTv2_500M: 0.69 (previous state-of-the-art Nucleotide Transformer v2 (500M)), Splice sites_MCC: 0.98, Promoters_MCC: 0.95, DNA_methylation_AUROC: 0.97 (HUES64), Promoter_strength_tobacco_PCC: 0.82, RNA_polyadenylation_PCC: 0.91 (ChatNT) vs 0.90 (APARENT2), Protein_melting_PCC: 0.89 (ChatNT) vs 0.85 (ESM2), RNA_degradation_PCC_human: 0.62 (ChatNT) vs 0.74 (Saluki), RNA_degradation_PCC_mouse: 0.63 (ChatNT) vs 0.71 (Saluki), Calibration_example: Examples predicted with probability 0.9 are correct ~90%; medium-confidence area less calibrated before Platt scaling, Overall_performance_preserved: Same performance (MCC) across tasks after deriving perplexity-based probabilities; calibration improved after Platt’s model |
| Application Domains | Genomics, Transcriptomics, Proteomics, Molecular biology / regulatory genomics, Computational biology / bioinformatics, Biomedical research (potential healthcare applications mentioned as extension) |
272. Predicting expression-altering promoter mutations with deep learning, Science (May 29, 2025)
| Category | Items |
|---|---|
| Datasets | GTEx v8 (Genotype-Tissue Expression), ENCODE bigWigs, FANTOM5 CAGE-seq bigWigs, Promoter variant training set (GTEx-derived), gnomAD r3.0, UK Biobank (UKBB) proteomics, Genomics England (GEL) 100,000 Genomes Project (aggV2/v17), Massively Parallel Reporter Assay (MPRA) eQTL dataset (published / generated), Promoter MPRA library (GEL-targeted), GENCODE v39 (human) and vM25 (mouse) TSS annotations, ClinVar (clinvar_20240819 VCF) |
| Models | Convolutional Neural Network, Feedforward Neural Network, Multi-Layer Perceptron, Autoencoder |
| Tasks | Regression, Binary Classification, Multi-class Classification, Anomaly Detection, Feature Extraction, Clustering, Dimensionality Reduction |
| Learning Methods | Supervised Learning, Pre-training, Fine-Tuning, Ensemble Learning, Representation Learning, Batch Learning, Mini-Batch Learning, Gradient Descent, Backpropagation, End-to-End Learning |
| Performance Highlights | validation_profile_prediction_loss: not reported numerically in text (model selected by lowest validation loss), GTEx_outlier_classification_auROC_under_vs_over: 0.89, GTEx_outlier_classification_auROC_under_vs_null: 0.80, GTEx_outlier_classification_auROC_over_vs_null: 0.74, GTEx_fine-mapped_eQTLs_auROC_under_vs_over: 0.87, GTEx_fine-mapped_eQTLs_auROC_under_vs_null: 0.79, GTEx_fine-mapped_eQTLs_auROC_over_vs_null: 0.75, correlation_with_eQTL_effect_sizes_Pearson_r: 0.56 (p = 8.2e-36), MPRA_eQTL_correlation_Pearson_r: 0.63 (p = 6.1e-77), MPRA_eQTL_auROC_under_vs_over: 0.90, MPRA_eQTL_auROC_under_vs_null: 0.81, MPRA_eQTL_auROC_over_vs_null: 0.83, UKBB_promoter_pQTLs_correlation_Pearson_r: 0.56 (p = 7.9e-10) for 104 cis-pQTLs not in LD; correlation increased to 0.60 for genes with high mRNA stability, UKBB_rare_promoter_variants_proteomics_correlation_Pearson_r: 0.48 (p = 2.8e-101), UKBB_proteomics_auROC_under_vs_over: 0.91, UKBB_proteomics_auROC_under_vs_null: 0.77, UKBB_proteomics_auROC_over_vs_null: 0.78, GEL_RNAseq_correlation_Pearson_r: 0.61 (p = 6.6e-171), GEL_RNAseq_auROC_under_vs_over: 0.90, GEL_RNAseq_auROC_under_vs_null: 0.78, GEL_RNAseq_auROC_over_vs_null: 0.78, ClinVar_auROC_pathogenic_vs_benign: 0.76, trans_expression_correction: model used to reduce trans-regulatory confounding to increase detection of multitissue outliers (quantitative improvement reported as increases from 2540 to 4030 outliers after progressive corrections), contextual_result_counts: 2540 outliers after PC correction -> 3116 after cis-eQTL correction -> 4030 after trans-expression correction, OUTRIDER_comparison: PromoterAI outperformed outlier detection based on OUTRIDER autoencoder in number/enrichment of multitissue outliers when matching false discovery rate in shuffled background (Fig. 1E) |
| Application Domains | Disease Genomics, Rare Disease Diagnosis, Functional Genomics, Population Genetics, Clinical Genetics / Diagnostic Interpretation, Proteomics / Biomarker discovery, Regulatory genomics / promoter biology, Computational genomics / variant effect prediction |
271. Probabilistic phase labeling and lattice refinement for autonomous materials research, npj Computational Materials (May 24, 2025)
| Category | Items |
|---|---|
| Datasets | Synthetic Ta-Sn-O calibration datasets (3 noise levels), Al-Li-Fe-O synthetic benchmark, CrxFe0.5−xVO4 experimental XRD set, High-quality Ca5(PO4)3F XRD spectrum, Ta-Sn-O experimental lg-LSA high-throughput dataset, XCA calibration dataset (single-phase synthetic patterns) |
| Models | Convolutional Neural Network, Non-negative Matrix Factorization, Gaussian Process |
| Tasks | Multi-label Classification, Classification, Regression, Dimensionality Reduction, Feature Extraction |
| Learning Methods | Supervised Learning, Expectation-Maximization, Ensemble Learning, Maximum A Posteriori, Backpropagation |
| Performance Highlights | ECE_low_noise: 9.81%, ECE_high_noise: 9.12%, training_set_sizes: models trained on 24,000 and 40,000 simulated spectra (synthetic benchmark); other training sizes used for experimental tests: 20k, 32.5k, 52k, reduction_example: 201 XRD patterns reduced to 4 basis patterns in the lg-LSA Ta-Sn-O example, speed_impact: enabled phase labeling on 4 NMF bases in 9 s (probabilistic labeling) and extension to all 201 spectra in 4 s on a 4-core M1 MacBook Air, background_modeling: Background modeled by kernel regressor with Matern kernel; jointly optimized with phase model to prevent overfitting, contextual_performance: Joint optimization prevents the background from overfitting, leading to physically meaningful decompositions (Fig. 2a). |
| Application Domains | Materials science (X-ray diffraction, crystallography), High-throughput experimentation (autonomous materials discovery), Spectral de-mixing and analysis (deterministic spectroscopy) |
270. Data-Driven Design of Mechanically Hard Soft Magnetic High-Entropy Alloys, Advanced Science (May 22, 2025)
| Category | Items |
|---|---|
| Datasets | HTP-DFT HEA database (this work) |
| Models | Random Forest, Gradient Boosting Tree, Multi-Layer Perceptron, Decision Tree |
| Tasks | Binary Classification, Regression, Dimensionality Reduction, Feature Selection |
| Learning Methods | Ensemble Learning, Supervised Learning, Feature Learning, Dimensionality Reduction, Gradient Boosting |
| Performance Highlights | accuracy: >90% (overall, training and test datasets); test accuracy ≈6% lower than training, R2_train_B0: 97.4%, R2_test_B0: 92.1%, R2_Mtot_train: >98%, R2_Mtot_test: >98%, R2_TC_train: 98.7%, R2_TC_test: ≈94.3% (test set performance 4.4% lower than training) |
| Application Domains | materials science, high-entropy alloys (HEAs), magnetic materials / soft magnets, mechanical property prediction (bulk modulus, alloy strength), computational materials design / high-throughput materials discovery |
269. A novel training-free approach to efficiently extracting material microstructures via visual large model, Acta Materialia (May 15, 2025)
| Category | Items |
|---|---|
| Datasets | PI-1 (pure iron), PI-2 (pure iron), SS (stainless steel), HEA (high-entropy alloy), LCS (low carbon steel) [11], DP590-1 (dual-phase steel), DP590-2 (dual-phase steel), NBS-1 (Ni-based superalloy) [16], NBS-2 (Ni-based superalloy) [16], AZA (AlZn alloy) [25], UHCS (ultrahigh carbon steel) [11] |
| Models | Vision Transformer, Multi-Layer Perceptron, Self-Attention Network, Cross-Attention, Attention Mechanism |
| Tasks | Semantic Segmentation, Instance Segmentation, Clustering |
| Learning Methods | Pre-training, Zero-Shot Learning, Prompt Learning, Transfer Learning, Fine-Tuning |
| Performance Highlights | PI-1 ARI (MatSAM): 0.62 (±2.4%), PI-1 F1 (MatSAM): 0.71 (±1.6%), SS ARI (MatSAM): 0.56 (±6.6%), PI-2 ARI (MatSAM): 0.75 (±2.0%), LCS ARI (MatSAM): 0.96 (±3.7%), DP590-1 IoU (MatSAM): 0.77 (±8.2%), DP590-2 IoU (MatSAM): 0.82 (±3.1%), NBS-1 IoU (MatSAM): 0.91 (±1.2%), NBS-2 IoU (MatSAM): 0.82 (±6.3%), AZA IoU (MatSAM): 0.96 (±0.6%), UHCS IoU (MatSAM): 0.76 (±0.7%), Average relative improvement vs best rule-based (ARI+IoU): 35.4% (reported average relative improvement combining ARI and IoU over best-performing conventional rule-based methods), Average improvement vs original SAM: 13.9% (reported average improvement over original SAM), Average IoU improvement vs specialist DL models (on 4 public datasets): 7.5% (reported average improvement), SAM baseline PI-1 ARI: 0.48 (±0.8%), SAM baseline PI-2 ARI: 0.69 (±3.3%), SAM baseline DP590-2 IoU: 0.68 (±6.7%), SAM baseline NBS-1 IoU: 0.85 (±7.6%), SAM baseline AZA IoU: 0.73 (±6.8%), LCS ARI (MatSAM vs OTSU vs Canny vs SAM): MatSAM 0.96 (±3.7%), SAM 0.86 (±9.8%), Canny 0.73 (±5.3%), OTSU 0.64 (±4.3%), NBS-1 inference time (MatSAM): 1812.67 ms per image (IoU 0.91), LCS inference time (MatSAM): 1926.35 ms per image (ARI 0.96) |
| Application Domains | Materials science, Materials microstructure analysis / characterization, Microscopy image analysis (OM, SEM, TEM, XCT), Automated quantitative microstructural characterization |
268. Interpretable Machine Learning Applications: A Promising Prospect of AI for Materials, Advanced Functional Materials (May 13, 2025)
| Category | Items |
|---|---|
| Datasets | 3.3 million materials science article abstracts (Tshitoyan et al.), SteelBERT pre-training corpus, Haeckelite candidate pool, Zhong et al. catalytic surfaces dataset, GNoME discovered stable-structure set, Polysulfone candidate set (dielectric polymer screening), DFT / first-principles datasets for ML interatomic potentials (examples) |
| Models | Linear Model, Polynomial Model, Decision Tree, Support Vector Machine, Gaussian Process, Gradient Boosting Tree, Ensemble Learning, Feedforward Neural Network, Multi-Layer Perceptron, Convolutional Neural Network, Variational Autoencoder, Generative Adversarial Network, Diffusion Model, Transformer, BERT, GPT, Graph Neural Network, ResNet |
| Tasks | Regression, Classification, Image Generation, Image Classification, Feature Selection, Feature Extraction, Clustering, Dimensionality Reduction, Language Modeling, Text Classification, Optimization, Image-to-Image Translation, Representation Learning |
| Learning Methods | Active Learning, Transfer Learning, Pre-training, Fine-Tuning, Adversarial Training, Self-Supervised Learning, Multi-Task Learning, In-Context Learning, Prompt Learning, Representation Learning, Ensemble Learning, Active Learning |
| Performance Highlights | discovered_structures: over 2.2 million potentially stable structures, search_space: explore over 1e60 possible compounds (conceptual capability), image_fidelity: high-fidelity synthetic images closely resembling experimental images, predicted_bandgap: identified hybrid perovskite composition with bandgap = 1.39 eV, classification_accuracy: >90%, example_designs: compositions with UTS of 600–950 MPa and electrical conductivity of 50.0% IACS |
| Application Domains | Materials science (general), Metallic structural materials (alloy design, high-entropy alloys), High-temperature alloys / superalloys, Battery materials and solid electrolytes, Perovskite photovoltaic materials, Catalytic materials and electrocatalysis (CO2 reduction, OER), Polymers (dielectric, high-thermal-conductivity polymers), Microstructure-informed manufacturing (additive manufacturing / LPBF), Protein structure prediction / biomolecular materials (AlphaFold examples), Drug-like molecule generation (TamGen), Optoelectronic and ferroelectric materials, Glass and ceramic materials, Porous materials and MOFs, Composite materials |
267. Exploration of crystal chemical space using text-guided generative artificial intelligence, Nature Communications (May 12, 2025)
| Category | Items |
|---|---|
| Datasets | Materials Project (MP-40 dataset, filtered <=40 atoms), MP-20 dataset, MatTPUSciBERT pretraining corpus, Generated TiO2 polymorphs (from Chemeleon), Generated Ti-Zn-O candidates, Generated Li-P-S-Cl candidates (quaternary), Test set (chronological split) |
| Models | Denoising Diffusion Probabilistic Model, Graph Neural Network, BERT, Transformer, Variational Autoencoder |
| Tasks | Structured Prediction, Graph Generation, Synthetic Data Generation, Distribution Estimation |
| Learning Methods | Contrastive Learning, Pre-training, Self-Supervised Learning, Generative Learning, Fine-Tuning |
| Performance Highlights | Validity_composition_prompt_BaselineBERT: 0.99, Uniqueness_composition_prompt_BaselineBERT: 0.94, StructureMatching_composition_prompt_BaselineBERT: 0.13, Metastability_composition_prompt_BaselineBERT: 0.22, Validity_formatted_text_BaselineBERT: 0.99, Uniqueness_formatted_text_BaselineBERT: 0.97, StructureMatching_formatted_text_BaselineBERT: 0.09, Metastability_formatted_text_BaselineBERT: 0.21, Validity_general_text_BaselineBERT: 0.99, Uniqueness_general_text_BaselineBERT: 0.97, StructureMatching_general_text_BaselineBERT: 0.06, Metastability_general_text_BaselineBERT: 0.23, Validity_composition_prompt_CrystalCLIP: 0.99, Uniqueness_composition_prompt_CrystalCLIP: 0.90, StructureMatching_composition_prompt_CrystalCLIP: 0.20, Metastability_composition_prompt_CrystalCLIP: 0.25, Validity_formatted_text_CrystalCLIP: 0.98, Uniqueness_formatted_text_CrystalCLIP: 0.92, StructureMatching_formatted_text_CrystalCLIP: 0.17, Metastability_formatted_text_CrystalCLIP: 0.19, Validity_general_text_CrystalCLIP: 0.99, Uniqueness_general_text_CrystalCLIP: 0.90, StructureMatching_general_text_CrystalCLIP: 0.20, Metastability_general_text_CrystalCLIP: 0.25, StructureMatching_overall_test_CrystalCLIP: 0.20, CompositionMatchingRate_on_MP-20_Chemeleon: 67.52%, StructureMatchRate_relative: lower than DiffCSP and FlowMM (strict criterion: structure match only counted when composition also matches), RMSE_relative: worse than DiffCSP; similar to FlowMM (exact RMSE values not provided), TiO2_generated_count: 549 sampled polymorphs (539 converged with MACE-MP), TiO2_DFT_identified_metastable_count: 122 unique metastable TiO2 structures (DFT-refined), TiO2_new_spacegroups: 50 structures with space groups not previously observed in known TiO2 polymorphs, Ti-Zn-O_predicted_stable: 1 structure below convex hull, Ti-Zn-O_predicted_metastable: 58 metastable structures, Li-P-S-Cl_predicted_stable: 17 new stable structures proposed, Li-P-S-Cl_predicted_metastable: 435 metastable structures generated, Li-P-S-Cl_energy_distribution_within_0.15eV: ≈80% of sampled configurations within 0.15 eV/atom above convex hull, StructureGeneration_runtime_Li-P-S-Cl: ≈72 hours on a single A100 GPU (search of Li-P-S-Cl space), CompositionMatching_ratio_trend: composition-matching ratio declines with increasing number of atoms; Crystal CLIP outperforms Baseline BERT by up to ~3x in composition matching across atom counts (Figure 3a) |
| Application Domains | Materials science / inorganic crystals, Crystal structure prediction and generation, Computational materials discovery and high-throughput screening, Solid-state battery materials (Li-P-S-Cl electrolyte space), Polymorph exploration (e.g., TiO2 polymorphs), Phase diagram construction |
266. Using GNN property predictors as molecule generators, Nature Communications (May 08, 2025)
| Category | Items |
|---|---|
| Datasets | QM9, ZINC subset (250,000 molecules), Generated DFT dataset (this work), QM9 (used as random draw / baseline) |
| Models | Graph Neural Network, Graph Convolutional Network, Graph Isomorphism Network, Graph Attention Network, GraphSAGE, Message Passing Neural Network, Variational Autoencoder, Normalizing Flow, Diffusion Model, Graph Convolutional Policy Network, CrippenNet, Genetic Algorithm (evolutionary baseline) |
| Tasks | Regression, Synthetic Data Generation, Optimization, Representation Learning |
| Learning Methods | Supervised Learning, Gradient Descent, Backpropagation, Mini-Batch Learning, Evolutionary Learning, Reinforcement Learning, Active Learning |
| Performance Highlights | test_MAE_QM9: 0.12 eV, generated_MAE_approx: 0.8 eV (observed on generated molecules vs DFT), ncalcs_per_target: 100 (per target in experiments), n_within_±0.5eV_for_4.1eV: 46 / 100, MAE_to_target_for_4.1eV: 0.81 eV, Diversity_for_4.1eV: 0.91 (average pairwise Tanimoto distance), n_within_±0.5eV_for_6.8eV: 50 / 100, MAE_to_target_for_6.8eV: 0.83 eV, Diversity_for_6.8eV: 0.90, n_within_±0.5eV_for_9.3eV: 34 / 100, MAE_to_target_for_9.3eV: 0.83 eV, Diversity_for_9.3eV: 0.83, Pearson_correlation_ρ_between_ML_and_DFT_all300: 0.86, average_time_per_in-target_molecule: 12.0 s (4.1 eV), 2.1 s (6.8 eV), 10.4 s (9.3 eV) on 4-CPU 3.40 GHz machine, training_data: ZINC subset (250k) + QM9, used_as_proxy_for_generation: yes, success_rate_−2.5<=logP<=−2: 43.5% (Proxy evaluation), diversity_−2.5..−2: 0.932 (average pairwise Tanimoto distance), success_rate_5<=logP<=5.5: 14.4% (Proxy evaluation), diversity_5..5.5: 0.917, average_time_per_in-target_molecule: 5.6 s for −2.5 to −2; 3.4 s for 5 to 5.5 on 4-CPU 3.40GHz, MAE_on_QM9test: 0.048 eV, MAE_on_generated_molecules: 1.16 eV (worse on generated molecules), generation_performance: slightly worse than the authors’ simple GNN used in main DIDgen experiments (no exact numbers in main text; details in SI), JANUS_DFT_ncalcs_for_4.1eV: 197, JANUS_DFT_n_within±0.5eV_for_4.1eV: 24 (12.2%), JANUS_DFT_MAE_for_4.1eV: 0.96 eV, JANUS_DFT_diversity_for_4.1eV: 0.79, JANUS_Proxy_for_4.1eV_MAE: 1.05 eV (proxy-run), comparison_note: DIDgen nearly matches or outperforms JANUS on the nine metrics reported in Table 1, success_rate_−2.5..−2(Proxy): 11.3%, diversity−2.5..−2: 0.846, success_rate_5..5.5(Proxy): 7.6%, diversity_5..5.5: 0.907, success_rate−2.5..−2(Oracle): 85.5%, diversity−2.5..−2: 0.392 (oracle-evaluated diversity low for that case), success_rate_5..5.5_(Oracle): 54.7%, diversity_5..5.5: 0.855 |
| Application Domains | Computational materials science, Molecular discovery / cheminformatics, Drug discovery (logP as proxy for cell permeability), Organic electronics / OLED materials (HOMO-LUMO gap targeting for emission wavelength), Automated experimentation / autonomous labs (context and motivation) |
265. Discovery of Sustainable Energy Materials Via the Machine-Learned Material Space, Small (May 05, 2025)
| Category | Items |
|---|---|
| Datasets | OptiMate dataset (Tr[Im(ϵ_ij)]/3 calculated with 300 meV broadening), Alexandria database (referenced) |
| Models | Graph Attention Network, Multi-Layer Perceptron, Message Passing Neural Network |
| Tasks | Regression, Dimensionality Reduction, Clustering, Feature Extraction |
| Learning Methods | Supervised Learning, Representation Learning, Feature Learning |
| Performance Highlights | None |
| Application Domains | Materials science, Optical materials, Energy materials, Photovoltaics (PV) and multijunction solar cells, Solar hydrogen generation, Optical sensors, Epsilon-near-zero materials, Energy-efficient light-emitting devices |
264. End-to-end data-driven weather prediction, Nature (May 2025)
| Category | Items |
|---|---|
| Datasets | ERA5 reanalysis, HadISD (Hadley Centre integrated surface dataset), ICOADS (International Comprehensive Ocean-Atmosphere Data Set), IGRA (Integrated Global Radiosonde Archive), ASCAT (Metop Advanced Scatterometer) Level 1B, AMSU-A / AMSU-B / Microwave Humidity Sounder / HIRS, IASI (Infrared Atmospheric Sounding Interferometer), GridSat (Gridded Geostationary Brightness Temperature Data), HRES (ECMWF Integrated Forecasting System high-resolution) forecasts, GFS (NCEP Global Forecast System) forecasts, NDFD (National Digital Forecast Database) |
| Models | Vision Transformer, U-Net, Multi-Layer Perceptron, Convolutional Neural Network, Encoder-Decoder, Multi-Head Attention, Self-Attention Network |
| Tasks | Time Series Forecasting, Regression, Image-to-Image Translation, Feature Extraction |
| Learning Methods | Supervised Learning, Pre-training, Fine-Tuning, End-to-End Learning, Transfer Learning, Stochastic Gradient Descent, Representation Learning |
| Performance Highlights | LW-RMSE: Aardvark achieved lower latitude-weighted RMSE than GFS across most lead times for many variables; approached HRES performance for most variables and lead times (held-out test year 2018, ERA5 ground truth), LW-RMSE at t=0: initial-state estimation error reported and compared to HRES analysis; Aardvark has non-zero error at t=0 against ERA5 whereas HRES also non-zero, MAE: Aardvark produced skilful station forecasts up to 10 days lead time; competitive with station-corrected HRES and matched NDFD over CONUS for 2-m temperature; for 10-m wind, mixed results (worse than station-corrected HRES over CONUS but outperformed NDFD)., Fine-tuning improvement (MAE %): 2-m temperature: −6% MAE (Europe, West Africa, Pacific, Global), −3% MAE (CONUS). 10-m wind speed: 1–2% MAE improvements for most regions (except Pacific)., Inference speed: Full forecast generation ~1 second on four NVIDIA A100 GPUs, Computational cost comparison: HRES data assimilation and forecasting ~1,000 node hours (operational NWP) |
| Application Domains | Numerical weather forecasting / atmospheric sciences, Local weather forecasting (station-level forecasts), Transportation (weather impacts), Agriculture (heatwaves, cold waves forecasting), Energy and renewable energy (wind forecasts), Public safety and emergency services (extreme weather warnings, tropical cyclones), Marine forecasting (ocean/ship observations), Insurance and finance (weather risk modelling), Environmental monitoring (potential extension to atmospheric chemistry and air quality), Operational meteorology (replacement/augmentation of NWP pipelines) |
263. Engineering principles for self-driving laboratories, Nature Chemical Engineering (May 2025)
| Category | Items |
|---|---|
| Datasets | None |
| Models | None |
| Tasks | Optimization, Experimental Design, Data Generation, Resource Allocation |
| Learning Methods | Reinforcement Learning, Active Learning, Online Learning |
| Performance Highlights | material_consumption_ratio_vs_conventional: less than 1/500, data_generation_rate_equivalent: over 100 researchers, quality_improvement: achieved superior optical properties compared with literature protocols, experiments_reduction: substantially reduces the number of experiments required (qualitative / orders-of-magnitude claims), timeline_reduction: reduce discovery and development timelines by orders of magnitude (qualitative) |
| Application Domains | Chemical engineering, Materials discovery, Pharmaceutical process development, Synthesis of (bio)molecules, Colloidal atomic layer deposition / nanostructure synthesis, Automated laboratory operations / self-driving laboratories (SDLs) |
262. Large language model-driven database for thermoelectric materials, Computational Materials Science (May 01, 2025)
| Category | Items |
|---|---|
| Datasets | Northeast Thermoelectric Materials Database (this work), Corpus of collected DOIs (Elsevier), Validation sample of extracted entries, Sierepeklis & Cole automated thermoelectric database, Gaultois et al. thermoelectric dataset (reference), Na and Chang thermoelectric database (reference), Various first-principles / ab initio thermoelectric databases (references: JARVIS, ab initio transport DB, etc.) |
| Models | GPT, Transformer, Graph Neural Network, Attention Mechanism |
| Tasks | Information Retrieval, Feature Extraction, Data Generation, Regression, Information Retrieval |
| Learning Methods | Prompt Learning, Supervised Learning, Unsupervised Learning, Pre-training |
| Performance Highlights | Composition_accuracy_percent: 90, Type_accuracy_percent: 91, Seebeck_Coefficient_accuracy_percent: 100, Seebeck_Obs_Temp_accuracy_percent: 100, Electrical_Conductivity_accuracy_percent: 100, Electrical_Conductivity_Obs_Temp_accuracy_percent: 100, Thermal_Conductivity_accuracy_percent: 100, Thermal_Conductivity_Obs_Temp_accuracy_percent: 100, Power_Factor_accuracy_percent: 100, Power_Factor_Obs_Temp_accuracy_percent: 100, ZT_accuracy_percent: 100, ZT_Obs_Temp_accuracy_percent: 100, Crystal_Structure_accuracy_percent: 97, Lattice_Structure_accuracy_percent: 96, Lattice_Parameters_accuracy_percent: 100, Space_Group_accuracy_percent: 100, Experimental_flag_accuracy_percent: 100 |
| Application Domains | Thermoelectric materials, Materials science, Energy harvesting, Sustainable energy / thermoelectric device design, Scientific text mining / literature curation |
261. Leveraging generative models with periodicity-aware, invertible and invariant representations for crystalline materials design, Nature Computational Science (May 2025)
| Category | Items |
|---|---|
| Datasets | Materials Project (MP), NOMAD, AFLOW, Vanadium–Oxygen dataset (example), CrystaLLM training set (standardized tokenized CIFs), MatterSim DFT training data (referenced), Open Catalyst Project (OC22) |
| Models | Generative Adversarial Network, Variational Autoencoder, Diffusion Model, Denoising Diffusion Probabilistic Model, Graph Neural Network, Graph Convolutional Network, Graph Attention Network, Transformer, Convolutional Neural Network, Message Passing Neural Network, Normalizing Flow |
| Tasks | Synthetic Data Generation, Graph Generation, Regression, Classification, Property Prediction (mapped to Regression/Classification), Optimization |
| Learning Methods | Generative Learning, Adversarial Training, Unsupervised Learning, Pre-training, Transfer Learning, Fine-Tuning, Active Learning, Domain Adaptation, Incremental Learning, Ensemble Learning, Representation Learning, Reinforcement Learning |
| Performance Highlights | local_energy_minimum_proximity: >15x closer (relative measure), samples_generated: 10,000, space_groups_sampled: 113, predicted_formation_energy: ~3.1 eV per atom (reported as ‘low’), pretraining_dataset_size: 2.3 million structures |
| Application Domains | crystalline materials design / inorganic crystals, battery materials, catalytic materials, superconductors, photoanodes, thermoelectric materials (power factor prediction), magnetic materials (low-supply-chain-risk magnets), electrolyte and cathode materials (high-temperature operation), optical/optoelectronic materials (e.g., n-type doped low-dimensional materials) |
260. Automated processing and transfer of two-dimensional materials with robotics, Nature Chemical Engineering (May 2025)
| Category | Items |
|---|---|
| Datasets | Transferred 4-inch graphene wafer electrical mapping (97 devices), Array of transferred graphene wafers (production runs), Transferred 2-inch MoS2 domains on 4-inch graphene wafer (heterostructure mapping), Surface and morphology characterization dataset (OM/AFM/Raman/PL/XPS/SEM/TEM data), Life cycle inventory (LCA) datasets and results, Automation control code (spin-coating and lamination/delamination machines) |
| Models | None |
| Tasks | Control, Planning, Optimization, Decision Making, Experimental Design, Resource Allocation |
| Learning Methods | None |
| Performance Highlights | None |
| Application Domains | 2D materials manufacturing, Electronics (device fabrication), Photonics, Quantum technology, Materials characterization and heterostructure fabrication, Industrial automation and robotics, Environmental assessment / life cycle analysis |
259. Self-driving nanoparticle synthesis, Nature Chemical Engineering (May 2025)
| Category | Items |
|---|---|
| Datasets | spectroscopic properties extracted from existing literature, AFION online extinction spectra (experimental data), Offline TEM and energy-dispersive X-ray spectroscopy (EDX) validation data |
| Models | Gaussian Process |
| Tasks | Optimization, Experimental Design, Hyperparameter Optimization, Regression |
| Learning Methods | Supervised Learning, Active Learning, Online Learning |
| Performance Highlights | experiments_to_identify_optimum: fewer than 30, time_to_identify_optimum: 30 hours, distinct_np_types_synthesized: 8, training_data_size: fewer than 30 experimental runs (as above) |
| Application Domains | Nanoparticle synthesis, Chemical engineering, Materials research, Autonomous experimentation / self-driving laboratories, Microfluidics-based synthesis and inline spectroscopy |
258. MatterChat: A Multi-Modal LLM for Material Science, Preprint (April 26, 2025)
| Category | Items |
|---|---|
| Datasets | Materials Project Trajectory (MPtrj) (relaxed samples), GNoME (newly discovered materials set used for comparative evaluation) |
| Models | Graph Neural Network, Transformer, Attention Mechanism, Multi-Head Attention |
| Tasks | Classification, Regression, Question Answering, Text Generation, Dimensionality Reduction, Clustering, Representation Learning |
| Learning Methods | Contrastive Learning, Supervised Learning, Fine-Tuning, Pre-training, Transfer Learning, Representation Learning, Distributed Learning |
| Performance Highlights | Accuracy: 0.6373, Accuracy: 0.6864, Accuracy: 0.8683, Accuracy: 0.8873, Accuracy: 0.8629, Accuracy: 0.7839, Accuracy: 0.8753, Accuracy: 0.8797, Accuracy: 0.7418, Accuracy: 0.7944, Accuracy: 0.8515, Accuracy: 0.8573, Accuracy: 0.7171, Accuracy: 0.6549, Accuracy: 0.8504, Accuracy: 0.857, Accuracy: 0.8339, Accuracy: 0.6833, Accuracy: 0.9368, Accuracy: 0.9333, Accuracy: 0.7759, Accuracy: 0.4238, Accuracy: 0.857, Accuracy: 0.8535, RMSE (eV/atom): 0.4105, RMSE (eV/atom): 1.8059, RMSE (eV/atom): 0.15, RMSE (eV/atom): 0.1212, RMSE (eV/atom): 0.4415, RMSE (eV/atom): 0.4051, RMSE (eV/atom): 0.1053, RMSE (eV/atom): 0.0964, RMSE (eV): 1.2516, RMSE (eV): 1.4725, RMSE (eV): 0.559, RMSE (eV): 0.5058 |
| Application Domains | Materials science (inorganic materials), Energy (materials for energy applications), Electronics (semiconductor materials), Catalysis (materials discovery for catalysis), Scientific human-AI interaction (material synthesis guidance and reasoning) |
257. Towards AI-driven autonomous growth of 2D materials based on a graphene case study, Communications Physics (April 25, 2025)
| Category | Items |
|---|---|
| Datasets | Raman spectroscopy measurements (experimentally collected spectra used for scoring), Atomic Force Microscopy (AFM) adhesion force maps, X-ray Photoelectron Spectroscopy (XPS) core-level spectra (C1s), Angle-Resolved Photoemission Spectroscopy (ARPES) intensity maps and MDCs |
| Models | Multi-Layer Perceptron, Gaussian Process |
| Tasks | Optimization, Control, Experimental Design |
| Learning Methods | Evolutionary Learning, Active Learning, Backpropagation, Gradient Descent |
| Performance Highlights | AFM_graphene_area_protocol1: 22.4%, AFM_graphene_area_protocol5: 88.2%, XPS_graphene_area_PTC1: 18.94%, XPS_graphene_area_PTC2: 19.60%, XPS_graphene_area_PTC4: 31.30%, XPS_graphene_area_PTC5: 23.10%, ARPES_MDC_FWHM_PTC1: 0.044 Å^-1, ARPES_MDC_FWHM_PTC5: 0.020 Å^-1, experiments_to_learn: “a few tens of experiments” (stated in text), one_experiment_per_learning_step: yes |
| Application Domains | Graphene growth / 2D materials synthesis, Materials science (epitaxial growth on SiC), Autonomous laboratories and experimental optimization, Surface characterization and electronic structure validation (Raman, AFM, XPS, ARPES) |
256. Science acceleration and accessibility with self-driving labs, Nature Communications (April 24, 2025)
| Category | Items |
|---|---|
| Datasets | Galaxy Zoo, Foldit, The Harvard Clean Energy Project |
| Models | Gaussian Process, Transformer, GPT, Multi-Layer Perceptron, Random Forest |
| Tasks | Optimization, Experimental Design, Image Classification, Ranking, Data Generation, Hyperparameter Optimization |
| Learning Methods | Active Learning, Reinforcement Learning, Ensemble Learning, Pre-training, Transfer Learning, Ensemble Learning |
| Performance Highlights | None |
| Application Domains | Chemical sciences / Chemistry, Materials science, Energy (e.g., photovoltaics, battery materials), Medicine / Pharmaceutical discovery, Nanoparticles and nanomaterials synthesis, Microscopy / Scanning probe and electron microscopy, Analytical method development (e.g., HPLC), Industrial R&D and specialty chemicals |
255. Learning Smooth and Expressive Interatomic Potentials for Physical Property Prediction, Preprint (April 23, 2025)
| Category | Items |
|---|---|
| Datasets | MPTrj, SPICE-MACE-OFF, OMat24 (O M A T 24), Matbench-Discovery, MDR Phonon benchmark, TM23, MD22, sAlex (subsampled Alexandria), SPICE (SPICE-1.0), PubChem / DES370K / Dipeptides / Sol. AA / Water / QMugs |
| Models | Message Passing Neural Network, Graph Neural Network, Transformer |
| Tasks | Binary Classification, Regression, Regression |
| Learning Methods | Supervised Learning, Pre-training, Fine-Tuning, Self-Supervised Learning, Transfer Learning, Backpropagation, Batch Learning |
| Performance Highlights | F1: 0.831, κSRME: 0.34, MAE (energy, eV/atom): 0.033, RMSD: 0.0752, Accuracy: 0.946, Precision: 0.804, R2: 0.822, F1: 0.925, κSRME: 0.17, MAE (energy, eV/atom): 0.018, RMSD: 0.0608, Accuracy: 0.977, Precision: 0.928, R2: 0.866, MAE(ωmax) (K): 21, MAE(S) (J/K/mol): 13, MAE(F) (kJ/mol): 5, MAE(CV) (J/K/mol): 4, Energy MAE (meV/atom) - MPTrj test: 17.02, Force MAE (meV/Å) - MPTrj test: 43.96, Stress MAE (meV/Å/atom) - MPTrj test: 0.14, Energy MAE (meV/atom) - SPICE test: 0.23, Force MAE (meV/Å) - SPICE test: 6.36, SPICE-MACE-OFF test splits (eSEN-6.5M) Energy MAE (meV/atom): {‘PubChem’: 0.15, ‘DES370K M.’: 0.13, ‘DES370K D.’: 0.15, ‘Dipeptides’: 0.07, ‘Sol. AA’: 0.25, ‘Water’: 0.15, ‘QMugs’: 0.12}, SPICE-MACE-OFF test splits (eSEN-6.5M) Force MAE (meV/Å): {‘PubChem’: 4.21, ‘DES370K M.’: 1.24, ‘DES370K D.’: 2.12, ‘Dipeptides’: 2.0, ‘Sol. AA’: 3.68, ‘Water’: 2.5, ‘QMugs’: 3.78}, Training efficiency reduction (wallclock): 40% reduction (conservative fine-tuned model vs from-scratch conservative trained for equivalent validation loss), Validation loss convergence: Conservative fine-tuned model achieves lower validation loss after 40 epochs compared to from-scratch conservative model trained for 100 epochs (Figure 3). |
| Application Domains | Materials science (inorganic materials, crystal stability prediction), Computational chemistry (molecular forces, energies for organic molecules and peptides), Molecular dynamics simulations (MD energy conservation and simulation stability), Phonon and vibrational property prediction (thermal conductivity, vibrational entropy, free energy, heat capacity), Drug discovery / biomolecular modeling (molecular datasets, peptides), Benchmarking and methodology for ML interatomic potentials (MLIPs) |
254. Harnessing database-supported high-throughput screening for the design of stable interlayers in halide-based all-solid-state batteries, Nature Communications (April 17, 2025)
| Category | Items |
|---|---|
| Datasets | Materials Project database (retrieved 21,576 Li-containing materials), Li3OCl experimental/characterization data (this work; Supplementary Data), DFT simulation data (interface models, DOS, reaction energies) — Supplementary Data 8 & 9, OMat24 inorganic materials dataset, MatterSim (deep learning atomistic model) (ref. 45) |
| Models | Transformer, Graph Neural Network, None (Density Functional Theory / DFT — not in provided model list) |
| Tasks | Ranking, Binary Classification, Regression, Recommendation, Text Generation |
| Learning Methods | Prompt Learning, Supervised Learning, High-throughput Screening (not in provided list) |
| Performance Highlights | None |
| Application Domains | All-solid-state Li metal batteries (ASSLMBs), Materials discovery for interlayer materials in halide solid-state electrolytes, Computational materials science (first-principles DFT + database screening), Electrochemistry / battery interface engineering, Scientific writing assisted by large language models (manuscript language editing) |
253. Data-driven discovery of biaxially strained single atoms array for hydrogen production, Nature Communications (April 17, 2025)
| Category | Items |
|---|---|
| Datasets | HT-DFT SAA Au-bMX2 hydrogen adsorption dataset (ΔGH*) |
| Models | Random Forest |
| Tasks | Regression, Feature Selection, Feature Extraction, Ranking, Classification |
| Learning Methods | Supervised Learning, Ensemble Learning, Bagging, Feature Selection, Cross-Validation |
| Performance Highlights | R2_test: 0.992, RMSE_test: 0.077, R2: 0.953, R2: 0.972, top_four_features_contribution_model_N: ≈70%, top_four_features_contribution_model_F: >76%, εads_importance_model_N: 53.98%, key_feature_identified: Electron affinity (EA) identified as most drastic influence on ΔGH* via SHAP/PFI |
| Application Domains | Electrocatalysis (Hydrogen Evolution Reaction), Computational materials discovery, Data-driven catalyst screening, Single-atom catalysis on transition metal dichalcogenides (TMDs) |
252. A Multiagent-Driven Robotic AI Chemist Enabling Autonomous Chemical Research On Demand, Journal of the American Chemical Society (April 16, 2025)
| Category | Items |
|---|---|
| Datasets | Literature Database (local), Protocol Library, Model Library (pretrained models), MO-HEC experimental dataset (random sampling), MO-HEC combined measured set (including optimized sample), Task 1 FTIR dataset, Task 2 PXRD dataset, Task 3 PQD fluorescence dataset, Task 4 g-C3N4 factorial experiment data, Task 5 BiOX photocatalytic degradation data, Task 7 photoreduction GC-MS timecourse data |
| Models | Transformer, Multi-Layer Perceptron, Dimensionality Reduction, Unsupervised Learning |
| Tasks | Information Retrieval, Feature Extraction, Regression, Optimization, Dimensionality Reduction, Feature Extraction, Information Retrieval |
| Learning Methods | Pre-training, Fine-Tuning, Transfer Learning, Supervised Learning, Unsupervised Learning, Prompt Learning, In-Context Learning |
| Performance Highlights | best_experimental_overpotential_at_10_mA_cm^-2_mV: 266.1, random_sampling_overpotentials_all_above_mV: 300+, stability_reduction_over_500_h_percent: <2%, HER_performance_range_mmol_g^-1: 9.28e-5 to 2.10e-3 |
| Application Domains | chemistry, materials science, electrocatalysis (oxygen evolution reaction), photocatalysis, organic synthesis / photocatalytic organic reactions, laboratory automation / robotics, autonomous self-driving laboratories (SDLs) |
251. Generative deep learning for predicting ultrahigh lattice thermal conductivity materials, npj Computational Materials (April 11, 2025)
| Category | Items |
|---|---|
| Datasets | AIRSS carbon periodic structures (Pickard), Generated CDVAE structures (this work), GAP-2020 subset (training set for pre-trained Allegro), Active-learning MLIP dataset (this work), Benchmarks (selected for detailed κL evaluation) |
| Models | Variational Autoencoder, Diffusion Model, Message Passing Neural Network, Multi-Layer Perceptron, Autoencoder |
| Tasks | Synthetic Data Generation, Regression, Clustering, Dimensionality Reduction, Feature Extraction, Representation Learning, Data Generation |
| Learning Methods | Generative Learning, Active Learning, Supervised Learning, Representation Learning, Ensemble Learning, Pre-training, Fine-Tuning, Gradient Descent, Active Learning |
| Performance Highlights | generated_structures: 100,000, generation_speed_per_structure: 0.48 seconds per structure (single RTX 2080 Ti GPU), unique_after_deduplication: 7,213 (≈7.2% of generated), candidates_after_symmetry_filter: 1,361, see: same as CDVAE generation performance above (100k generated, 7213 unique, 1361 candidates), energy_MAE_on_test: 24.3 meV atom^-1, force_RMSE_on_test: 273 meV Å^-1, uncertainty_threshold: 15 meV atom^-1, benchmarks_with_κL_over_800: 9 of 53 benchmarks confirmed, total_identified_ultrahigh_κL_structures: 34 structures with κL > 800 W m^-1K^-1, max_κL_found: up to 2,400 W m^-1K^-1 (aside from diamond), component_role: used inside Allegro (2-body latent MLP: [128,128,128]; edge energy MLP: [256,128,64]), global_model_performance: see Allegro metrics above (energy MAE 24.3 meV atom^-1, force RMSE 273 meV Å^-1), latent_space_used_for: sampling new structures; facilitates global optimization in latent space (conceptual), benchmarks_selected: 50 most diverse via FPS + 5 reported = 53 benchmarks, KNN_selected_unique_structures: 64 unique structures (from KNN clusters), KNN_high_κL_rate: Over 50% of KNN-selected materials exhibit κL > 800 W m^-1K^-1 |
| Application Domains | Materials discovery / computational materials science, Thermal management and heat transport (lattice thermal conductivity prediction), Crystal structure prediction, Carbon materials / carbon allotropes, Atomistic simulations and interatomic potential development, High-throughput virtual screening of materials |
250. Electronic Structure Guided Inverse Design Using Generative Models, Preprint (April 08, 2025)
| Category | Items |
|---|---|
| Datasets | MP DOS (curated by this paper) |
| Models | Denoising Diffusion Probabilistic Model, Graph Neural Network, Variational Autoencoder, Generative Adversarial Network, Normalizing Flow |
| Tasks | Synthetic Data Generation, Graph Generation, Regression, Data Generation |
| Learning Methods | Generative Learning, Self-Supervised Learning, Supervised Learning |
| Performance Highlights | Classifier-free MAE(ŷgen, y) T=200: 0.102, Classifier-free MAE(ŷgen,ŷ) T=200: 0.049, Classifier-free MAE(ŷgen, y) T=500: 0.109, Classifier-free MAE(ŷgen,ŷ) T=500: 0.065, Classifier-free MAE(ŷgen, y) T=1000: 0.120, Classifier-free MAE(ŷgen,ŷ) T=1000: 0.088, Structure match % (Classifier-free) T=200: 81.6, Composition match % (Classifier-free) T=200: 94.4, Structure match % (Classifier-free) T=500: 42.0, Composition match % (Classifier-free) T=500: 73.0, Structure match % (Classifier-free) T=1000: 14.7, Composition match % (Classifier-free) T=1000: 58.7, Generated set size for large-scale screening: 10,000, Post-filtering candidates (formation energy ≤ -1.5 eV/atom): 108, Selected for DFT validation: 8, Classifier MAE(ŷgen, y) T=200: 0.114, Classifier MAE(ŷgen,ŷ) T=200: 0.068, Classifier MAE(ŷgen, y) T=500: 0.200, Classifier MAE(ŷgen,ŷ) T=500: 0.182, Classifier MAE(ŷgen, y) T=1000: 0.268, Classifier MAE(ŷgen,ŷ) T=1000: 0.256, Structure match % (Classifier) T=200: 63.5, Composition match % (Classifier) T=200: 73.8, Structure match % (Classifier) T=500: 4.23 (some exclusions), Composition match % (Classifier) T=500: 6.01 (some exclusions), Structure match % (Classifier) T=1000: 0.07, Composition match % (Classifier) T=1000: 2.95, Failures mapping to surrogate at T=1000 (Classifier): 314 out of 10,158, Surrogate forward model MAE(ŷ, y): 0.096 |
| Application Domains | materials discovery / inverse materials design, electronic structure prediction and design (density of states conditioned design), catalysis (design of catalytic materials), photovoltaics, superconductors, computational materials science (high-throughput screening with MLFF and DFT validation) |
249. Leveraging data mining, active learning, and domain adaptation for efficient discovery of advanced oxygen evolution electrocatalysts, Science Advances (April 04, 2025)
| Category | Items |
|---|---|
| Datasets | Domain-knowledge literature dataset (full), Domain-knowledge literature dataset (high-quality subset), Active-learning experimental dataset (DASH experimental runs), High-throughput DFT dataset (source domain S), DFT dataset (target domain T) |
| Models | Support Vector Machine, Gradient Boosting Tree, XGBoost, LightGBM, CatBoost, Random Forest, Decision Tree, XGBoost, Multi-Layer Perceptron, Convolutional Neural Network, Recurrent Neural Network, Long Short-Term Memory, Gated Recurrent Unit, Bidirectional LSTM, Temporal Convolutional Network |
| Tasks | Regression, Dimensionality Reduction, Feature Selection, Feature Extraction, Experimental Design, Optimization, Hyperparameter Optimization |
| Learning Methods | Unsupervised Learning, Supervised Learning, Active Learning, Domain Adaptation, Transfer Learning, Fine-Tuning, Ensemble Learning, Boosting |
| Performance Highlights | R2: 0.84, MAE: 29.76 mV (for η10 on full dataset), MAE: 27.21 mV (for η10 on high-quality dataset), R2 (stability, full dataset): 0.86, R2: 0.89 (stability on high-quality dataset), R2: close to or over 0.99 on Dataset S (source domain), R2_on_T: substantially improved over Committee T (exact numeric improvement not specified), R2_on_S_post-adaptation: primarily in range 0.8 to 0.9 (retained predictive power on S), best_η10_over_iterations: decreased from 209 mV to 154 mV, failure_rate_first_batch: 47%, failure_rate_fourth_and_fifth_batches: 0%, total_experimental_samples: 258 samples over five iterations, DFT_theoretical_overpotential_for_sample_C: 376 mV in commonly studied descriptor scenario, stability_descriptors_for_sample_C: Udiss = 3.34 V; ΔGVO = 3.84 eV, experimental_decay_rate_for_sample_C: 0.1728 and 0.1964 mV hour−1 at 10 and 20 mA cm−2 over 125 hours |
| Application Domains | Materials Science, Electrocatalysis, Acidic Oxygen Evolution Reaction (OER), Proton Exchange Membrane (PEM) Water Electrolysis, Computational Materials Science / DFT surrogate modeling |
248. A high-throughput experimentation platform for data-driven discovery in electrochemistry, Science Advances (April 04, 2025)
| Category | Items |
|---|---|
| Datasets | LCE dataset of electrolyte formulations (final), Library of 180 small-molecule additives, DFT descriptors dataset (quantum chemistry descriptors for selected additives), Coin cell cycling validation dataset (reservoir half-cell protocol), Reproducibility CE measurements (platform benchmarking) |
| Models | Linear Model, Support Vector Machine, Gaussian Process, Random Forest, Gradient Boosting Tree, XGBoost, Multi-Layer Perceptron |
| Tasks | Regression, Feature Selection, Feature Extraction, Data Generation |
| Learning Methods | Supervised Learning, Ensemble Learning, Stacking, Feature Selection |
| Performance Highlights | R2: 0.86, RMSE: 0.142, MAE: 0.104, R2: 0.81, RMSE: 0.165, MAE: 0.112, R2: 0.32, RMSE: 0.310, MAE: 0.251, RMSE_percent_CE: 2.074%, MAE_percent_CE: 1.768%, Predicted_CE_percent: 98.02%, Predicted_LCE: 1.703, Experimental_average_CE_percent_over_200_cycles: 99.52% |
| Application Domains | Electrochemistry, Aqueous zinc metal batteries (AZMBs) / Battery research, Energy storage and conversion, Automated high-throughput experimentation (HTE) for materials discovery, Materials science (electrode/electrolyte optimization, electroplating), Electrocatalysis / electro-organic synthesis / corrosion studies (noted as broader applicability) |
247. Physics-informed, dual-objective optimization of high-entropy-alloy nanozymes by a robotic AI chemist, Matter (April 02, 2025)
| Category | Items |
|---|---|
| Datasets | Web of Science peroxidase abstracts (literature + patents), MD / composition dataset (simulated HEA compositions), DFT surface-structure dataset, Experimental HEA dataset (synthesized and measured), ML training split for thermodynamic NN models |
| Models | Gaussian Process, Feedforward Neural Network, Multi-Layer Perceptron, XGBoost, GPT |
| Tasks | Optimization, Regression, Binary Classification, Clustering, Dimensionality Reduction, Feature Extraction |
| Learning Methods | Supervised Learning, Unsupervised Learning, Boosting, Ensemble Learning, In-Context Learning |
| Performance Highlights | best_Vmax/KM_PI-DO-BO_step12_s^-1: 2.973e-3, best_Vmax/KM_PI-DO-BO_step12_s^-1_alt: 2.19e-3, kcat/KM (derived) M^-1 s^-1: 4.41e7 and 3.25e7, baseline_random_highest_Vmax/KM_s^-1: 6.18e-5, standard_DO-BO_highest_Vmax/KM_s^-1: 2.86e-5, step6_highest_Vmax/KM_s^-1: <1.03e-4, steps7-10_highest_Vmax/KM_s^-1: 1.58e-3, natural_HRP_kcat/KM_M^-1 s^-1 (reported literature): 9.42e5, training_dataset_size: 12,205 compositions (80% train / 20% test), classification_target: DG–OH_des class 0/1 (threshold: -1 eV), impact_on_search_efficiency: Not provided as single numeric ML metric; reflected by improvement in discovered catalytic efficiencies (Vmax/KM increased from <1e-4 to 2.973e-3 after GPT-in-the-loop introduction), calibration_model_accuracy: reported as high predictive accuracies for formulation→composition NN (see Figures S7 F–S7O), no numeric test error in main text |
| Application Domains | materials science, catalysis, nanozymes / enzymatic mimics, automated materials discovery / robotic experimentation, computational materials chemistry (DFT/MD + ML integration), chemical synthesis optimization |
246. Towards multimodal foundation models in molecular cell biology, Nature (April 2025)
| Category | Items |
|---|---|
| Datasets | Human Cell Atlas (HCA), Human Biomolecular Atlas Program (HuBMAP), Human Tumor Atlas Network (HTAN), ENCODE, International Human Epigenome Consortium (IHEC), CellxGENE aggregated collection (including HCA and HuBMAP), Perturb-seq / large-scale CRISPR perturbation datasets, Protocol-specific paired-modal datasets (10x Multiome, CITE-seq, ASAP-seq, TEA-seq etc.), Reference pretraining collections cited (examples used to illustrate scale) |
| Models | Transformer, GPT, BERT, Swin Transformer, Autoencoder, Graph Neural Network |
| Tasks | Clustering, Dimensionality Reduction, Classification, Regression, Sequence-to-Sequence, Time Series Forecasting, Data Generation, Synthetic Data Generation, Graph Generation |
| Learning Methods | Self-Supervised Learning, Pre-training, Transfer Learning, Fine-Tuning, In-Context Learning, Contrastive Learning, Supervised Learning, Prompt Learning, Active Learning |
| Performance Highlights | None |
| Application Domains | Molecular cell biology, Genomics, Transcriptomics (single-cell RNA-seq), Epigenomics (chromatin accessibility, methylation), Proteomics, Metabolomics, Spatial profiling / spatial transcriptomics, Drug discovery and perturbation response prediction, Biomarker discovery, Personalized medicine / clinical cohort analysis |
245. Applications of natural language processing and large language models in materials discovery, npj Computational Materials (March 24, 2025)
| Category | Items |
|---|---|
| Datasets | Materials-related abstracts (Tshitoyan corpus), Materials-related abstracts (Pei corpus), ChemDataExtractor auto-generated datasets (perovskite, dye-sensitized, band gaps, etc.), 800 hand-annotated NER corpus (Weston et al.), Polymer annotated abstracts (Shetty et al.), Superalloy dataset (Wang et al.), Superalloy article corpus (processing/synthesis actions), Solid-state synthesis recipes knowledge base, Large article corpus for oxide synthesis extraction (Kim et al.), Perovskite solar cell dataset (Xie et al.), TransPolymer pretraining data (PI1M augmented), polyBERT training data (hypothetical polymers), Steel corpus for SteelBERT, Domain-specific corpora: MatSciBERT / SCIBERT / MaterialBERT / BatteryBERT / OpticalBERT |
| Models | Transformer, BERT, GPT, Attention Mechanism, Self-Attention Network, Seq2Seq, Bidirectional LSTM, Conditional Random Field, Latent Dirichlet Allocation, Variational Autoencoder |
| Tasks | Information Retrieval, Named Entity Recognition, Relation Extraction, Text Classification, Sequence-to-Sequence, Clustering, Question Answering, Information Retrieval, Regression, Data Generation, Structured Prediction, Sequence Labeling |
| Learning Methods | Self-Supervised Learning, Supervised Learning, Semi-Supervised Learning, Pre-training, Fine-Tuning, Prompt Learning, Few-Shot Learning, Zero-Shot Learning, Transfer Learning, Reinforcement Learning, Multi-Task Learning, Knowledge Distillation, Contrastive Learning, In-Context Learning |
| Performance Highlights | F1-score: 87%, extracted_records: ~300,000 polymer property records, precision: 90–99%, recall: 90–99%, F1-score: 90–99%, F1-score: up to 0.98, accuracy: 0.96, bulk_modulus_precision: 90.8%, bulk_modulus_recall: 87.7%, critical_cooling_precision: 91.6%, critical_cooling_recall: 83.6%, schema_generation_F1: 87.14%, R2_yield_strength: 78.17% (±3.40%), R2_ultimate_tensile_strength: 82.56% (±1.96%), R2_total_elongation: 81.44% (±2.98%), text_search_accuracy: 96.9%, property_prediction_accuracy: 95.7%, structure_generation_accuracy: 87.5%, MaScQA_improvement: up to 20.61%, SciQA_improvement: up to 45.73%, extraction_accuracy: 73% (parsing success) |
| Application Domains | Materials science (general), Alloys and superalloys, Polymers, Perovskite solar cells / photovoltaics, Metal-organic frameworks (MOFs), Catalysis (inorganic catalysts / binary alloy catalysts), Inorganic materials synthesis (solid-state, solution-based), Steels and metallurgy, Batteries (battery materials), Optical materials, Thermoelectrics, Metallic glasses, Cement and concrete, Two-dimensional materials, Autonomous chemical experiments / laboratory automation |
244. Elemental numerical descriptions to enhance classification and regression model performance for high-entropy alloys, npj Computational Materials (March 18, 2025)
| Category | Items |
|---|---|
| Datasets | HEA phase dataset (Al-Ti-V-Cr-Fe-Co-Ni-Cu), Hardness dataset for HEAs, Virtual composition space (sampling pool), Experimental validation set (synthesized HEAs A1-A15), Six additional materials datasets (tested for generality) |
| Models | Generalized Linear Model, Gradient Boosting Tree, Decision Tree, Random Forest, Naive Bayes, Multi-Layer Perceptron, Support Vector Machine |
| Tasks | Binary Classification, Multi-class Classification, Regression, Feature Extraction, Feature Selection |
| Learning Methods | Supervised Learning, Evolutionary Learning, Active Learning, Reinforcement Learning, Boosting, Ensemble Learning |
| Performance Highlights | accuracy: 0.87, accuracy: 0.97, MAE: 45.8 HV, R2: 0.88, R2: improved (values not explicitly listed in main text), MAPE: improved (values not explicitly listed in main text), accuracy_gain_range: 3% to 22%, experimental_validation: 13/15 correct for SS/NSS; 8/9 correct for FCC/BCC/DP |
| Application Domains | Materials science, High-entropy alloys (HEAs) phase prediction and property prediction, Mechanical property prediction (hardness, high-temperature strength, fracture strain), Functional ceramics (BaTiO3: piezoelectric d33, electrostrain, dielectric energy storage density), Shape memory alloys (NiTi-based transformation temperatures) |
242. Transformers and genome language models, Nature Machine Intelligence (March 2025)
| Category | Items |
|---|---|
| Datasets | GenBank, RefSeq, Sequence Read Archive (SRA), ENCODE, Roadmap Epigenomics, GTEx, 1000 Genomes Project, GenomicBenchmarks (GenomicBenchmarks / GenomicBenchmarks datasets), Genome Understanding Evaluation (GUE), BEND (Benchmarking DNA Language models), NCBI Genome (eight Brassicales reference genomes), Single reference human genome (one reference genome), Prokaryotic genomes (whole genomes), CRISPR perturbation screens (reference experiments) |
| Models | Transformer, BERT, GPT, Convolutional Neural Network, Recurrent Neural Network, Encoder-Decoder, Seq2Seq, Attention Mechanism, Self-Attention Network, Multi-Head Attention, State Space Model, U-Net |
| Tasks | Language Modeling, Self-Supervised Learning, Classification, Binary Classification, Regression, Structured Prediction, Sequence-to-Sequence, Language Modeling, Feature Extraction, Anomaly Detection |
| Learning Methods | Self-Supervised Learning, Pre-training, Fine-Tuning, Transfer Learning, Unsupervised Learning, Contrastive Learning, Few-Shot Learning, Zero-Shot Learning, Supervised Learning, Language Modeling, Representation Learning |
| Performance Highlights | accuracy_description: high accuracy reported (no numeric value provided in paper), comparative_performance: DNABERT-2 performs comparably to Nucleotide Transformer on several tasks despite 21x fewer parameters (no numeric metrics provided), zero_few_shot_rank: Nucleotide Transformer Multi-Species and original DNABERT had the best zero- and few-shot embeddings (qualitative, no numeric values given), benchmark_performance: HyenaDNA achieved ‘state-of-the-art performance on all eight datasets from GenomicBenchmarks’ (qualitative statement, no numeric values provided), comparative_performance: Hybrid models combining CNN + transformer (e.g., Enformer, Borzoi) show improved assay prediction and increased receptive field; exact numeric metrics not provided in review, efficiency_claim: Selective SSMs / Mamba and Hyena-like layers claim improved scaling (subquadratic) and competitive accuracy with lower compute cost (qualitative in paper), specific_task_performance: HyenaDNA reported strong performance at much larger context windows (1 million nucleotides) and strong benchmark results (see HyenaDNA entry)., generalization_claim: Models pretrained on inter-species (multi-species) data generalize better on human prediction tasks than intra-species (population-scale) pretraining (qualitative claim, no numeric metrics provided), qualitative: GPN (a convolutional/transformer-modified model) learned non-coding variant effects from unsupervised pretraining and outperformed supervised DeepSEA in reported comparisons (qualitative claim in review), embedding_distance_use: Cosine distances between reference and variant sequence embeddings used to indicate functional differences (method described; no numeric metrics given) |
| Application Domains | genomics / regulatory genomics, functional genomics (TF-binding, chromatin accessibility, histone marks), 3D genome architecture and chromatin contact mapping, variant effect prediction and interpretation, gene expression prediction (bulk and single-cell contexts), computational biology and network biology, synthetic biology (in silico perturbation and design), personalized medicine and clinical genomics, drug discovery (future direction), single-cell transcriptomics (scRNA-seq foundation models) |
241. The deep finite element method: A deep learning framework integrating the physics-informed neural networks with the finite element method, Computer Methods in Applied Mechanics and Engineering (March 01, 2025)
| Category | Items |
|---|---|
| Datasets | Plate with a circular hole (case study), Non-symmetric stretching plate (case study), Rock drill boom (3D case study), FEM (Abaqus) reference solutions |
| Models | Multi-Layer Perceptron, Feedforward Neural Network |
| Tasks | Regression, Optimization |
| Learning Methods | Pre-training, Fine-Tuning, Stochastic Gradient Descent, Gradient Descent, Backpropagation, Supervised Learning |
| Performance Highlights | plate_with_hole_relative_error_max_DFEM_selected_points: <= 0.16 %, plate_with_hole_relative_error_range_DEM_selected_points: 10.25 % - 16.71 %, plate_with_hole_max_abs_error_DFEM_u: 0.02, plate_with_hole_max_abs_error_DFEM_v: 0.015, plate_with_hole_max_abs_error_DEM_u: 0.2, plate_with_hole_max_abs_error_DEM_v: 0.08, training_time_reduction_near_pretraining_load: DFEM ~1/8 of FEM time (example), pretraining_convergence_epochs: 8-10 epochs (fine-tuning convergence reported), rock_drill_boom_elements: 46,337, rock_drill_boom_nodal_points: 10,205, rock_drill_boom_max_error_u: 2.5e-3, rock_drill_boom_max_error_v: 1.8e-3, rock_drill_boom_max_error_w: 1.3e-2, rock_drill_boom_avg_error_u: 3.2e-4, rock_drill_boom_avg_error_v: 4.6e-4, rock_drill_boom_avg_error_w: 2.8e-3, rock_drill_boom_relative_errors_selected_points_DFEM: <= 0.54 %, computation_time_reduction_with_pretraining_vs_FEM: ≈ 66% (example from Table 6), DFEM_relative_error_table3example_Point1: 0.62 %, DEM_relative_error_table3_example_Point1(2500iter): 14.68 %, DEM_relative_error_table3_example_Point1(5000_iter): 99.99 %, DEM_special_training_relative_error_Point1: 1.15 %, DFEM_error_reduction_vs_DEM: up to 99% reduction in relative error (reported across cases) |
| Application Domains | Solid elasticity mechanics, Computational mechanics, Three-dimensional structural analysis, Engineering structural analysis, Digital twin applications |
240. A generative model for inorganic materials design, Nature (March 2025)
| Category | Items |
|---|---|
| Datasets | Alex-MP-20, Alex-MP-ICSD, Materials Project (MP), Alexandria, Labelled magnetic density dataset, Labelled bandgap dataset, Labelled bulk modulus dataset, ICSD (test structures) |
| Models | Denoising Diffusion Probabilistic Model, Variational Autoencoder, Graph Neural Network, Feedforward Neural Network |
| Tasks | Synthetic Data Generation, Graph Generation, Regression, Optimization, Experimental Design |
| Learning Methods | Pre-training, Fine-Tuning, Supervised Learning, Unsupervised Learning, Generative Learning, Transfer Learning |
| Performance Highlights | energy_above_hull_below_0.1eV_MP: 78%, energy_above_hull_below_0_eV_MP: 13%, energy_above_hull_below_0.1_eV_Alex-MP-ICSD: 75%, energy_above_hull_below_0_eV_Alex-MP-ICSD: 3%, RMSD_below_0.076_A_for_generated_structures: 95% (of generated 1,024 structures), unique_at_1000_samples: 100%, unique_at_10_million_samples: 52%, new_structures_fraction: 61% (new vs Alex-MP-ICSD), SUN_increase_vs_CDVAE_and_DiffCSP: 60% more SUN structures, average_RMSD_reduction_vs_baselines: 50% lower RMSD, SUN_increase_over_MatterGen-MP: 70% increase, RMSD_decrease_over_MatterGen-MP: 5x decrease, overall_improvement_vs_prior_SOTA: more than 2x likelihood to be SUN; up to order-of-magnitude closer to local energy minimum, labelled_dataset_size: 605,000 DFT magnetic density labels, SUN_structures_with_mag_density>0.2_A^-3_using_180_DFT_calcs: up to 18, labelled_dataset_size: 42,000 DFT bandgap labels, target_bandgap: 3.0 eV, distribution_shift_towards_target: substantial shift in property distribution among SUN samples towards desired target (Fig. 4b), labelled_dataset_size: 5,000 DFT bulk modulus labels, SUN_found_with_budget_180_DFT_calcs: 106 SUN structures (95 distinct compositions), screening_baseline_with_budget_180_DFT_calcs: 40 SUN structures (28 distinct compositions), DFT_predicted_bulk_modulus_of_ordered_target_structure: 222 GPa, experimental_estimated_bulk_modulus: up to 169 GPa (best of four measurements); 158 ± 11 GPa reported, DFT_MAE_on_95_matches: 23 GPa, DFT_RMSE_on_95_matches: 32 GPa, samples_per_target_for_generation: 8,192 candidates per target bulk modulus value, generated_samples_needed_for_quinary_system_performance: 10,240 (MatterGen) vs ~70,000 (substitution) vs ~600,000 (RSS), higher_percentage_of_SUN_structures_across_system_types: MatterGen generates highest percentage of SUN structures for each system type and complexity (Fig. 3a,b), unique_structures_on_combined_convex_hull: MatterGen finds highest number in partially and well-explored systems (Fig. 3c) |
| Application Domains | Materials design (inorganic crystalline materials), Energy storage (materials discovery), Catalysis (materials discovery), Carbon capture (materials discovery), Permanent magnet discovery (magnetic materials), Superhard materials discovery (mechanical properties), Experimental materials synthesis and validation |
239. CrystalFlow: A Flow-Based Generative Model for Crystalline Materials, Preprint (February 24, 2025)
| Category | Items |
|---|---|
| Datasets | MP-20, MPTS-52, MP-CALYPSO-60 |
| Models | Normalizing Flow, Graph Neural Network, Multi-Layer Perceptron, Variational Autoencoder, Diffusion Model, Normalizing Flow, Transformer |
| Tasks | Data Generation, Synthetic Data Generation, Graph Generation, Optimization |
| Learning Methods | Supervised Learning, Backpropagation, Representation Learning, End-to-End Learning, Gradient Descent |
| Performance Highlights | MP-20 k=1 MR (%): 62.02, MP-20 k=1 RMSE: 0.0710, MP-20 k=20 MR (%): 78.34, MP-20 k=20 RMSE: 0.0577, MP-20 k=100 MR (%): 82.49, MP-20 k=100 RMSE: 0.0513, MPTS-52 k=1 MR (%): 22.71, MPTS-52 k=1 RMSE: 0.1548, MPTS-52 k=20 MR (%): 40.37, MPTS-52 k=20 RMSE: 0.1576, MPTS-52 k=100 MR (%): 52.14, MPTS-52 k=100 RMSE: 0.1603, MP-CALYPSO-60 (500 generated) convergence rate CR (%) Cond-CDVAE S=5000: 82.20, MP-CALYPSO-60 (500) ion-steps Cond-CDVAE S=5000: 45.91, MP-CALYPSO-60 (500) CR (%) CrystalFlow S=100: 89.20, MP-CALYPSO-60 (500) ion-steps CrystalFlow S=100: 49.84, MP-CALYPSO-60 (500) CR (%) CrystalFlow S=1000: 90.20, MP-CALYPSO-60 (500) ion-steps CrystalFlow S=1000: 39.40, MP-CALYPSO-60 (500) CR (%) CrystalFlow S=5000: 90.60, MP-CALYPSO-60 (500) ion-steps CrystalFlow S=5000: 39.82, SiO2 case (200 samples) CR (%) Cond-CDVAE S=5000: 96.00, SiO2 case (200) ion-steps Cond-CDVAE S=5000: 44.36, SiO2 case (200) CR (%) CrystalFlow S=100: 100.00, SiO2 case (200) ion-steps CrystalFlow S=100: 35.65, SiO2 case (200) CR (%) CrystalFlow S=1000: 100.00, SiO2 case (200) ion-steps CrystalFlow S=1000: 35.84, SiO2 case (200) CR (%) CrystalFlow S=5000: 100.00, SiO2 case (200) ion-steps CrystalFlow S=5000: 31.99, DNG: structural validity (%): 99.55, DNG: compositional validity (%): 81.96, DNG: coverage recall (%): 98.21, DNG: coverage precision (%): 99.84, DNG: wdist(density): 0.169 (smallest among compared), DNG: wdist(Nel): 0.259, Inference time (min/10k generated) CrystalFlow RTX 4090 S=100: 4.1, Inference time (min/10k) CrystalFlow RTX 4090 S=1000: 37.0, Baseline DiffCSP RTX 4090 S=1000: 44.7, FlowMM A800 S=750: 65.1, FlowLLM A800 S=250: 89.6, MP-20 k=1 MR (%) CDVAE: 33.90, MP-20 k=1 RMSE CDVAE: 0.1045, MPTS-52 k=1 MR (%) CDVAE: 5.34, MPTS-52 k=1 RMSE CDVAE: 0.2106, MP-20 k=1 MR (%) DiffCSP: 51.49, MP-20 k=1 RMSE DiffCSP: 0.0631, MPTS-52 k=1 MR (%) DiffCSP: 12.19, MPTS-52 k=1 RMSE DiffCSP: 0.1786, MP-20 k=20 MR (%) DiffCSP: 77.93, MP-20 k=20 RMSE DiffCSP: 0.0492, MPTS-52 k=20 MR (%) DiffCSP: 34.02, MPTS-52 k=20 RMSE DiffCSP: 0.1749, MP-20 k=1 MR (%) FlowMM: 61.39, MP-20 k=1 RMSE FlowMM: 0.0566, MPTS-52 k=1 MR (%) FlowMM: 17.54, MPTS-52 k=1 RMSE FlowMM: 0.1726 |
| Application Domains | Crystalline materials generation, Crystal structure prediction (CSP), Inverse materials design / de novo materials generation, High-pressure materials discovery (pressure-conditioned generation), Computational condensed matter physics and materials science |
238. Genome modeling and design across all domains of life with Evo 2, Preprint (February 21, 2025)
| Category | Items |
|---|---|
| Datasets | OpenGenome2, GTDB representative prokaryotic genomes, Eukaryotic reference genomes (NCBI), Metagenomic sequences (curated), Organelle genomes (NCBI Organelle), mRNA and ncRNA transcripts (GTF-derived), EPDnew promoter sequences, ClinVar (2024.02.28 release), SpliceVarDB, BRCA1 saturation mutagenesis dataset (Findlay et al. 2018), BRCA2 variant dataset (Huang et al., 2025), Deep Mutational Scanning (DMS) datasets (ProteinGym and other compendia), DEG (Database of Essential Genes) and phage essentiality screens, lncRNA essentiality screens (Liang et al., 2024), Woolly mammoth genome (Sandoval-Velasco et al., 2024), Mycoplasma genitalium reference genome (Gibson et al., 2008), Saccharomyces cerevisiae chromosome III |
| Models | Convolutional Neural Network, Attention Mechanism, Self-Attention Network, Multi-Head Attention, Autoencoder, Feedforward Neural Network, Multi-Layer Perceptron, Transformer |
| Tasks | Regression, Binary Classification, Sequence Labeling, Synthetic Data Generation, Feature Extraction, Optimization, Experimental Design, Classification |
| Learning Methods | Self-Supervised Learning, Zero-Shot Learning, Supervised Learning, Pre-training, Fine-Tuning, Representation Learning, Contrastive Learning, Ensemble Learning |
| Performance Highlights | Spearman_correlation_DMS: competitive with state-of-the-art autoregressive protein language models; state-of-the-art on noncoding RNA fitness prediction (no single aggregated numeric value reported in main text), ClinVar_noncoding_SNV_AUROC_Evo2_40B: 0.987, ClinVar_noncoding_SNV_AUPRC_Evo2_40B: 0.974, ClinVar_coding_SNV_AUROC_Evo2_40B: 0.841, ClinVar_coding_SNV_AUPRC_Evo2_40B: 0.889, SpliceVarDB_intronic_SNV_AUROC_Evo2_40B: 0.926, SpliceVarDB_intronic_SNV_AUPRC_Evo2_40B: 0.971, SpliceVarDB_exonic_SNV_AUROC_Evo2_40B: 0.684, SpliceVarDB_exonic_SNV_AUPRC_Evo2_40B: 0.523, BRCA1_supervised_test_AUROC: 0.95, BRCA1_supervised_test_AUPRC: 0.86, BRCA1_coding_SNV_test_AUROC: 0.94, BRCA1_coding_SNV_test_AUPRC: 0.84, Exon_classifier_AUROC_range: 0.82-0.99 across eight held-out species, SAE_features_identified: features corresponded to exon/intron boundaries, transcription factor motifs, protein secondary structure, prophage regions (qualitative mapping; numeric F1/precision/recall reported for certain features e.g., exon/intron features evaluated on 1,000 genes but aggregate values varied per feature), Designed_chromatin_AUROC: ≈0.9 for many patterns when sampling >=30 128-bp chunks and selecting top-2 chunks per step; inference-time scaling shows log-linear improvement with more sampled tokens, Token_sampling_examples: sampling 30 or more 128-bp chunks and selecting top 2 chunks per step sufficient to achieve AUROCs around 0.9, Mitochondrial_generation_gene_counts: Evo 2 generated mitochondria with the correct number of CDS, tRNA, and rRNA genes (annotation via MitoZ); BLASTp analyses show varied percent identity to natural proteins (see Table S6: many genes with high percent identity; examples: mt-Atp6 91.59% to Alcelaphus buselaphus; mt-Nd6 94.43% to Neolissochilus hendersoni), M_genitalium_Pfam_hit_rate: ∼70% of Prodigal-annotated Evo 2 40B genes had significant Pfam/HHpred hits (E-value < 0.001), versus Evo 1 131k at ∼18%, Yeast_generation_feature_presence: Generated yeast chromosomes contained predicted tRNAs, promoters, and intronic structure albeit at lower density than native genome (quantified distributions in Figures 5L and S8) |
| Application Domains | genomics (prokaryotic and eukaryotic), clinical genetics / variant interpretation, molecular biology (protein/RNA function prediction), synthetic biology / genome design, epigenomics (chromatin accessibility design), comparative genomics and paleogenomics, bioinformatics / genome annotation, protein structure prediction (downstream evaluation via AlphaFold/ESMFold) |
237. Accelerating crystal structure search through active learning with neural networks for rapid relaxations, npj Computational Materials (February 20, 2025)
| Category | Items |
|---|---|
| Datasets | Initial candidate pools (benchmark systems: Si16, Na8Cl8, Ga8As8, Al4O6), Transferability candidate pools (Si transfer / Al2O3 transfer experiments), Training datasets (iteratively labeled by DFT during active learning), MaterialsProject reference structures (target minima) |
| Models | Message Passing Neural Network, Multi-Layer Perceptron, Graph Neural Network, Gaussian Process |
| Tasks | Regression, Optimization, Clustering, Data Generation, Feature Extraction, Representation Learning |
| Learning Methods | Active Learning, Supervised Learning, Ensemble Learning, Unsupervised Learning, Transfer Learning, Representation Learning, Pre-training |
| Performance Highlights | single-point DFT evaluations until convergence (benchmarks): 400–700, reduction_in_DFT_evaluations: up to two orders of magnitude (compared to AIRSS, Bayesian optimization, LAQA baselines), avoided_DFT: up to 95% of demanding DFT calculations (statement in Discussion), median_validation_steps_per_structure: 10.5 steps, median_energy_difference_after_validation: 2.6 meV/atom, Si transferability_total_DFT_CPU_hours: transfer: ~4322 CPU hours vs baseline: ~8863 CPU hours (≈58% reduction), Al2O3 transferability_reduction: ≈74% reduction in computational cost for DFT calculations (CPU hours) for Al2O3 transferability experiments, DFT_evaluations_for_transfer_Si: ~500 single-point DFT evaluations until convergence (per transfer simulation, ~4–5 cycles), reported_issue: learning interatomic forces with graph neural networks on relaxation trajectories of crystal structures has shown poor performance (cited Ref.66) |
| Application Domains | crystal structure search, materials discovery / computational materials science, global optimization of crystal compositions, high-throughput virtual screening (HTVS) for materials |
236. Autonomous platform for solution processing of electronic polymers, Nature Communications (February 17, 2025)
| Category | Items |
|---|---|
| Datasets | PEDOT_experiment.csv (Polybot experimental dataset) |
| Models | Gaussian Process, Random Forest |
| Tasks | Regression, Regression, Optimization, Image Classification, Dimensionality Reduction, Feature Selection, Hyperparameter Optimization |
| Learning Methods | Supervised Learning, Active Learning, Ensemble Learning |
| Performance Highlights | training_data_average_conductivity_S/cm: 276, all_data_average_conductivity_S/cm: 664, scale-up_average_conductivity_S/cm: >4500, training_data_average_coverage_%: 48, all_data_average_coverage_%: 72, search_space_size_conditions: 933,120 possible experimental conditions, experiment_iterations_run: initial 30 LHS + iterations shown 1–45 (in manuscript figures) ; termination criteria: two-week budget or performance plateau |
| Application Domains | Materials science, Electronic polymers (PEDOT:PSS), Thin-film processing and manufacturing, Printable electronics / transparent conductive films, Autonomous laboratories / self-driving lab platforms |
235. Developing novel low-density high-entropy superalloys with high strength and superior creep resistance guided by automated machine learning, Acta Materialia (February 15, 2025)
| Category | Items |
|---|---|
| Datasets | Curated Ni-Fe-Co-Al-Ti-Nb-Ta-Cr-Mo-W superalloy database (domain-knowledge preprocessed) |
| Models | Gradient Boosting Tree |
| Tasks | Regression, Optimization, Feature Extraction, Hyperparameter Optimization |
| Learning Methods | Supervised Learning, Ensemble Learning, Stacking, Evolutionary Learning, Feature Selection |
| Performance Highlights | R2_training_σYS-RT: 98%, RMSE_training_σYS-RT: 33~35 MPa, R2_training_σYS-HT: 96%, RMSE_training_σYS-HT: 40~45 MPa, Reported_overall: R2 ≥ 95% and RMSE ≤ 50 MPa (summary in Conclusions), AutoML_summary: AutoGluon autotuning and ensembling produced high predictive accuracy (see R2/RMSE above), population_generated: 46,380,000 candidate compositions generated when sampling wide composition ranges (reported as possible population), GA_iterations: GA terminated after 100 iterations; convergence observed after ~30 iterations in example, candidate_reduction_example: Without cluster-formula constraint, 156 candidate compositions correspond to σYSs = 1150 MPa; with cluster-formula constraint only R2 composition selected |
| Application Domains | Materials science, Alloy design / metallurgy, High-temperature structural materials (superalloys), Automated materials discovery (AI-guided composition design) |
234. Machine Learning in Solid-State Hydrogen Storage Materials: Challenges and Perspectives, Advanced Materials (February 12, 2025)
| Category | Items |
|---|---|
| Datasets | HydPARK, ML-HydPARK v0.0.0, ML-HydPARK v0.0.1, ML-HydPARK v0.0.2, ML-HydPARK v0.0.3, ML-HydPARK v0.0.4, ML-HydPARK v0.0.5, Complex and high-density HSMs proprietary dataset, AB2-type HSMs dataset (proprietary / curated), Mg-based HSMs (DFT data), Mg-based experimental dataset, Materials Project (MP), Crystallography Open Database (COD), ICSD / OQMD / NOMAD / AFLOWLIB / Pauling file / NIST, Proprietary LiBH4 catalyst dataset, Hypothetical ternary borohydrides dataset (CGCNN training), MaterialClouds 2D structure database (used for 2D MgH2 sheets GAN) |
| Models | Linear Model, Support Vector Machine, K-Nearest Neighbors, Decision Tree, Random Forest, Gradient Boosting Tree, XGBoost, Gaussian Process, Multi-Layer Perceptron, Feedforward Neural Network, Convolutional Neural Network, Graph Neural Network, Gaussian Process, Generative Adversarial Network, Deep Convolutional GAN, Denoising Diffusion Probabilistic Model, Crystal Graph Convolutional Neural Network, MEGNet (Materials Graph Network) — mapped to Graph Neural Network, DeePMD (Deep Potential Molecular Dynamics), SchNet (as part of SchNet-SSCHA), Ensemble models (stacking / bagging / boosting), Radial Basis Function Network |
| Tasks | Regression, Multi-class Classification, Clustering, Image Generation, Data Generation / Synthetic Data Generation, Clustering (k-means), Feature Selection / Dimensionality Reduction, Image Classification |
| Learning Methods | Supervised Learning, Unsupervised Learning, Transfer Learning, Semi-Supervised Learning, Active Learning, Ensemble Learning, Bagging, Boosting, Stacking, Evolutionary Learning, Pre-training / Fine-Tuning, Active/Automated Data Augmentation (GAN/VAE), Multi-Objective Optimization (Bayesian Optimization / MOBO) |
| Performance Highlights | MAE: 0.003 wt% H2, RMSE: 0.012 wt% H2, R2: 0.83, Accuracy: 0.8, Number_of_clusters: 3, Intra-cluster_cohesion: 0.6, Inter-cluster_separation: 1.4, MSE: 0.102 eV^2, MAE_train: 0.47, MAE_test: 1.52, MAE: 8.56 kJ mol^-1, MRE: 28%, HYST_R2: 0.81, HYST_MAE: 0.45 wt% H2, THOR_R2: 0.89, THOR_MAE: 4.53 kJ mol^-1 H2, Hydride_formation_enthalpy_R2: 0.647, Hydride_enthalpy_MAE: 4.36 kJ mol^-1 H2, Phase_abundance_R2: 0.832, Hydrogen_storage_capacity_R2: 0.688, Hydrogen_storage_capacity_MAE: 0.101 wt% H2, R2: 0.969, MRE: 2.291%, MSE: 3.909 kJ^2 mol^-2 H2, RMSE: 2.501 kJ mol^-1 H2, STD: 1.878 kJ mol^-1 H2, Best_model_R2: 0.980, STD: 0.043 wt% H2, MSE: 0.002 wt%2 H2, RMSE: 0.045 wt% H2, MAPE_max_discharge: 2.35%, RMSE_max_discharge: 9.74 mAh/g, R_max_discharge: 0.808, MAPE_fast_discharge: 0.89%, RMSE_fast_discharge: 1.38, R_fast_discharge: 0.991, MAE: 8.58 kJ mol^-1, RMSE: 11.73 kJ mol^-1, R2: 0.783, Pearson_Correlation: 0.885, H/M_MAE: 0.12, H/M_R2: 0.79, Enthalpy_MAE: 4.2 kJ mol^-1 H2 (R2=0.90), Entropy_MAE: 11 J mol^-1 K^-1 H2 (R2=0.69), LnPeq_MAE: 1.1 (R2=0.93), MAE: 3.1 meV/atom, ΔEmono_MAE: 0.04 eV (R2=0.71), ΔEdi_MAE: 0.04 eV (R2=0.93), Correlation_coefficient_R: >0.95, R2: 0.95, RMSE: 29.98 K, RMSE: 0.43 meV/atom, Energy_MAE: 0.02 eV, Force_MAE: 0.02 eV Å^-1, Structure_validation_ratio: 96.8%, Structure_generation_ratio: 87.3%, MSE: 1.144 wt%2 H2, RMSE: 1.066 wt% H2, EV: 0.889, R2: 0.888, Pearson_Correlation: 0.944, Spearman_Correlation: 0.949, MAE: 0.063 eV/atom, R2: >=0.96, Max_relative_error_absorption: < 8.0%, Max_relative_error_desorption: < 6.6%, Temperature-based_model_R2: 0.9798, Temperature-based_model_MAE: 0.046, Pressure-based_model_R2: 0.9946, Pressure-based_model_MAE: 0.00267, MAE: 5.5 kJ mol^-1 H2 |
| Application Domains | solid-state hydrogen storage materials (HSMs) research and design, high-throughput materials screening and discovery, thermodynamic and kinetic property prediction (formation energy, hydride enthalpy, equilibrium pressure, de-/hydrogenation temperature), design and optimization of hydrogen storage devices (hydride beds, reactors), interatomic potential development and molecular dynamics simulation (MLIPs for Mg-H systems), inverse materials design and multi-objective optimization for alloy selection, data augmentation and structure generation (2D hydride structure generation via GANs), electrochemical hydrogen storage (Ni-MH battery cathode performance prediction) |
232. ORGANA: A robotic assistant for automated chemistry experimentation and characterization, Matter (February 05, 2025)
| Category | Items |
|---|---|
| Datasets | ORGANA perception evaluation dataset, Electrochemistry experiment measurements (ORGANA runs), Electrochemistry experiment measurements (human chemists), Solubility / recrystallization / pH experiment measurements, User study data (chemists interacting with ORGANA) |
| Models | GPT, Transformer, Vision Transformer, CLIP, Seq2Seq |
| Tasks | Sequence-to-Sequence, Language Modeling, Zero-Shot Learning, Object Detection, Instance Segmentation, Pose Estimation, Regression, Image Classification, Sequence Labeling |
| Learning Methods | Prompt Learning, In-Context Learning, Zero-Shot Learning, Pre-training, Maximum Likelihood Estimation, Fine-Tuning |
| Performance Highlights | solubility_accuracy_vs_literature: salt 7±2%, sugar 11±2%, alum 12±3%, user_time_startup_written: 7.35 min, user_time_startup_spoken: 4.27 min, troubleshooting_time: 1.30 min, CLAIRify_style_time: 17.65 min, ORGANA_pKa1: 8.03 ± 0.17, chemists_pKa1: 8.02, ORGANA_slope: -61.4 ± 0.5 mV/pH unit, chemists_slope: -62.7 mV/pH unit, sequential_execution_time_avg: 21.67 min, parallel_execution_time_avg: 17.10 min, time_reduction: 21.1%, sequential_planning_time: 61.52 ± 0.1 s, temporal_TAMP_planning_time: 186.3 ± 46.0 s |
| Application Domains | chemistry lab automation, electrochemistry / flow battery characterization, materials discovery, robotics (manipulation and motion planning), robotic perception (transparent object perception), human-robot interaction / usability in lab settings |
231. Harnessing Large Language Models to Collect and Analyze Metal–Organic Framework Property Data Set, Journal of the American Chemical Society (February 05, 2025)
| Category | Items |
|---|---|
| Datasets | L2M3_Database (text-mined MOF dataset), Extracted synthesis-condition records, Density dataset (for regression experiments), Cambridge Structural Database (CSD) subset / CoREMOF references |
| Models | GPT, Transformer, Graph Convolutional Network, Random Forest, XGBoost, Support Vector Machine, K-nearest neighbor |
| Tasks | Information Retrieval, Classification, Structured Prediction, Regression, Recommendation, Question Answering, Dialog Generation, Entity Matching, Data Generation |
| Learning Methods | Fine-Tuning, Prompt Learning, Transfer Learning, Pre-training, Few-Shot Learning, Zero-Shot Learning, Supervised Learning |
| Performance Highlights | Categorization_Synthesis: {‘Precision’: 1.0, ‘Recall’: 0.98, ‘F1Score’: 0.99}, Inclusion_Synthesis: {‘Precision’: 0.96, ‘Recall’: 0.91, ‘F1Score’: 0.94}, Extraction_Synthesis: {‘Precision’: 0.96, ‘Recall’: 0.9, ‘F1Score’: 0.93}, Categorization_Property: {‘Precision’: 0.98, ‘Recall’: 0.94, ‘F1Score’: 0.96}, Inclusion_Property: {‘Precision’: 0.98, ‘Recall’: 0.98, ‘F1Score’: 0.98}, Extraction_Property: {‘Precision’: 0.97, ‘Recall’: 0.9, ‘F1Score’: 0.93}, Categorization_Table: {‘Precision’: 0.99, ‘Recall’: 1.0, ‘F1Score’: 1.0}, Inclusion_Table: {‘Precision’: 1.0, ‘Recall’: 1.0, ‘F1Score’: 1.0}, Extraction_Table: {‘Precision’: 1.0, ‘Recall’: 1.0, ‘F1Score’: 1.0}, R2_TrainExp_TestExp: 0.803, R2_TrainSim_TestExp: 0.495, R2_TrainExp_TestExp: 0.793, R2_TrainSim_TestExp: 0.469, R2_TrainExp_TestExp: 0.734, R2_TrainSim_TestExp: 0.415, R2_TrainExp_TestExp: 0.573, R2_TrainSim_TestExp: 0.151, R2_TrainExp_TestExp_CGCNN: 0.815, R2_TrainSim_TestExp_CGCNN: 0.4, R2_TrainExp_TestExp_MOFTransformer: 0.892, R2_TrainSim_TestExp_MOFTransformer: 0.38, RecommendationScore_FineTuned_GPT-4o_median: 0.83, RecommendationScore_FineTuned_GPT-3.5-turbo_median: 0.83, RecommendationScore_ZeroShot: approx. random (low), RecommendationScore_FewShot_n=100: exceeds statistical method but below fine-tuned models |
| Application Domains | Materials Science, Metal−Organic Frameworks (MOFs), Chemistry (synthetic chemistry / synthesis planning), Crystallography (structure-property linking), Scientific text mining / Natural Language Processing for scientific literature |
230. Automating the practice of science: Opportunities, challenges, and implications, Proceedings of the National Academy of Sciences (February 04, 2025)
| Category | Items |
|---|---|
| Datasets | Open Quantum Materials Database (OQMD), Materials Project (materials genome), Materials databases (general; stable materials databases referenced), DANDI, OpenNeuro, DABI, BossDB, BIDS (Brain Imaging Data Structure) - community standard, Open Science Framework, Large gene databases (general reference), Amazon Mechanical Turk (subject pool/platform), Prolific (prolific.ac) (subject pool/platform), JDRF-CGM trial data (Juvenile Diabetes Research Foundation continuous glucose monitoring trial), Elicit training data (LLM trained on paper abstracts) - referenced tool |
| Models | Transformer, GPT, Attention Mechanism, Autoencoder, Variational Autoencoder, Gaussian Process, Multi-Layer Perceptron, Autoencoder (again - reduced-order modeling) , Neural Architecture Search |
| Tasks | Regression, Dimensionality Reduction, Language Modeling, Text Summarization, Text Generation, Information Retrieval, Optimization |
| Learning Methods | Active Learning, Reinforcement Learning, Transfer Learning, Fine-Tuning, Pre-training, Representation Learning, Self-Supervised Learning, Neural Architecture Search |
| Performance Highlights | qualitative: BrainGPT “demonstrated the capability to outperform human experts in predicting the results of neuroscience experiments”, cost_per_article_usd: 15 |
| Application Domains | Materials science, Chemistry, Functional genomics / biology, Drug discovery, Behavioral sciences / psychology / cognitive science, Neuroscience, Physics (including plasma physics, fluid dynamics), Engineering (automation, robotics), Clinical health (e.g., diabetes monitoring) |
229. Exploration of Chemical Space Through Automated Reasoning, Angewandte Chemie (February 03, 2025)
| Category | Items |
|---|---|
| Datasets | Liverpool ionic conductivity dataset, Compositions generated by Comgen (this work) |
| Models | Diffusion Model |
| Tasks | Clustering, Classification, Data Generation, Synthetic Data Generation |
| Learning Methods | Unsupervised Learning, Supervised Learning, Generative Learning |
| Performance Highlights | predicted_high_conductivity_count_>1e-4S_cm^-1: 9 candidates, candidates_with_energy_from_convex_hull<=45meV_atom^-1: 8 candidates, selected_high_conductivity_reference_subset_count>=1e-3_S_cm^-1: 55 compounds (from Liverpool dataset) used as reference set for Mg-analogue search |
| Application Domains | Materials discovery, Materials chemistry, Solid-state battery electrolytes (Li-ion, Mg-ion conductors), Crystal structure prediction / computational materials design, Automated scientific reasoning / computational design workflows |
228. From text to insight: large language models for chemical data extraction, Chemical Society Reviews (February 03, 2025)
| Category | Items |
|---|---|
| Datasets | EuroPMC, arXiv, ChemRxiv, S2ORC (Semantic Scholar Open Research Corpus), Elsevier OA CC-BY Corpus, Open Reaction Database (ORD), USPTO, SciBERT pretraining corpus, MatSciBERT pretraining corpus, Llama 3 public data (mentioned) |
| Models | Transformer, GPT, BERT, Vision Transformer, Attention Mechanism, Self-Attention Network, Multi-Head Attention, Recurrent Neural Network |
| Tasks | Named Entity Recognition, Relation Extraction, Sequence Labeling, Information Retrieval, Text Classification, Image Classification, Question Answering, Clustering, Image-to-Image Translation |
| Learning Methods | Zero-Shot Learning, Few-Shot Learning, One-Shot Learning, In-Context Learning, Prompt Learning, Fine-Tuning, Pre-training, Supervised Learning, Self-Supervised Learning, Reinforcement Learning, Transfer Learning, Multi-Task Learning, Few-Shot Learning |
| Performance Highlights | annotation_time_reduction: >50% (time per sample reduced by more than half for last annotated abstracts, as reported) |
| Application Domains | Chemistry, Materials science, Organic synthesis, Inorganic materials and metal–organic frameworks (MOFs), Nanoparticles / nanomaterials, Polymers, Catalysis, Battery and energy materials, Spectroscopy / NMR data extraction, Scientific publishing / literature mining |
227. Balancing autonomy and expertise in autonomous synthesis laboratories, Nature Computational Science (February 2025)
| Category | Items |
|---|---|
| Datasets | generic computational datasets, low-cost and fast proxy measurement datasets, generic experimental datasets, specialized, standardized, and carefully evaluated datasets, simulated characterization data |
| Models | None |
| Tasks | Information Retrieval, Feature Extraction, Experimental Design, Data Augmentation, Anomaly Detection, Regression, Binary Classification |
| Learning Methods | Pre-training, Fine-Tuning, Transfer Learning, Active Learning, Supervised Learning, Domain Adaptation, Representation Learning |
| Performance Highlights | None |
| Application Domains | autonomous synthesis laboratories, materials synthesis, chemical synthesis, materials characterization, laboratory automation and robotics, experimental chemistry |
226. Knowledge-guided large language model for material science, Review of Materials Research (February 01, 2025)
| Category | Items |
|---|---|
| Datasets | Common Crawl, The Pile, StarCoder, Hugging Face Datasets, LIMA dataset, arxiv-physics-instruct (arxiv_instructed_Physics), peS2o, Gutenberg Project, PRM800K, Materials Project, OQMD (Open Quantum Materials Database), OMat24 (Open Materials 2024), OCx24 (Open Catalyst Experiments 2024), MoLFormer training data |
| Models | Transformer, Recurrent Neural Network, BERT, GPT, Attention Mechanism, Graph Neural Network, Diffusion Model, Sequence-to-Sequence |
| Tasks | Information Retrieval, Named Entity Recognition, Structured Prediction, Regression, Graph Generation, Language Modeling, Sequence-to-Sequence, Experimental Design, Planning, Data Generation, Anomaly Detection |
| Learning Methods | Fine-Tuning, Supervised Learning, Reinforcement Learning, Pre-training, In-Context Learning, Prompt Learning, Active Learning, Self-Supervised Learning, Transfer Learning |
| Performance Highlights | search_accuracy: 96.9%, property_prediction_accuracy: 95.7%, novelty_stability: >2x (more than twice as novel and stable), local_energy_proximity: 15x (15 times closer to the local energy minimum), benchmark_outperformance: outperforms graph-based and supervised models across ten molecular property prediction benchmarks, training_efficiency: requires 60x fewer GPUs for training, relative_metrics: outperform BERT-based methods in precision, recall, and F1 (no numeric values given), precision: ≈90%, recall: ≈90%, experiments_run: 355, success_rate_targets: 71% (41 of 58 targets), throughput: over two new materials per day, synthesis_success_examples: synthesized DEET, three thiourea organo-catalysts, and a novel chromophore (qualitative successes), validity_uniqueness_novelty: reported as high validity, uniqueness, and novelty (no numeric values provided) |
| Application Domains | Materials science (materials informatics, inorganic materials, metal-organic frameworks, polymers, catalysts), Chemistry / computational chemistry (molecular generation, reaction planning), Autonomous laboratories and robotics (experimental synthesis automation), Scientific text mining and structured information extraction, Drug discovery / molecular property prediction (mentioned as analogous domain) |
225. Battery lifetime prediction across diverse ageing conditions with inter-cell deep learning, Nature Machine Intelligence (February 2025)
| Category | Items |
|---|---|
| Datasets | MATR (MATR-1, MATR-2), HUST, CLO, CALCE, HNEI, UL-PUR, RWTH, SNL, MIX (MIX-100 and MIX-20), LFP subset (rich-resource chemistry), LCO subset (target chemistry in transfer experiments), NCA subset (target chemistry in transfer experiments), NMC subset (target chemistry in transfer experiments) |
| Models | Convolutional Neural Network, Multi-Layer Perceptron, Long Short-Term Memory, Random Forest, Support Vector Machine, Linear Model |
| Tasks | Regression, Time Series Forecasting, Transfer Learning |
| Learning Methods | Supervised Learning, Multi-Task Learning, Pre-training, Fine-Tuning, Representation Learning, Transfer Learning |
| Performance Highlights | r.m.s.e.reduction_vs_best_baseline%MATR-1: 36.5, r.m.s.e._reduction_vs_best_baseline%MATR-2: 6.8, r.m.s.e._reduction_vs_best_baseline%HUST: 20.1, r.m.s.e._reduction_vs_best_baseline%MIX-100: 27.4, r.m.s.e._reduction_vs_best_baseline%MIX-20: 40.1, MAPE_reduction_vs_single-cell_CNN%_average: up to 40 |
| Application Domains | Lithium-ion battery lifetime prediction / battery degradation modeling, Battery state-of-charge and state-of-health estimation (potential application mentioned), Cross-chemistry transfer for battery materials (LFP → LCO/NCA/NMC) and low-resource chemistries, Potential extension to fast-charging protocols and emerging chemistries (solid-state, sodium-ion) |
224. A guidance to intelligent metamaterials and metamaterials intelligence, Nature Communications (January 29, 2025)
| Category | Items |
|---|---|
| Datasets | None |
| Models | Multi-Layer Perceptron, Convolutional Neural Network, Recurrent Neural Network, Variational Autoencoder, Generative Adversarial Network, Diffusion Model, Graph Neural Network, Autoencoder, Encoder-Decoder, Bayesian Network |
| Tasks | Regression, Image-to-Image Translation, Image Classification, Image Generation, Clustering, Dimensionality Reduction, Feature Extraction |
| Learning Methods | Supervised Learning, Unsupervised Learning, Semi-Supervised Learning, Transfer Learning, Reinforcement Learning, Backpropagation, Pre-training, Fine-Tuning, Ensemble Learning |
| Performance Highlights | accuracy: 87%, relative_error_reduction: 23%, steps_to_solution: 9000, training_time: 8 hours for a five-layer diffractive ONN with 0.2M neurons, compute_reduction: two to three orders of magnitude reduction in required computation (example), spectral_similarity: 99.8% (in KK-driven causal neural network example), average_element_error: 1e-4, solved_equations: 8 complex equations demonstrated |
| Application Domains | Metamaterials / Metasurfaces design, Photonics / Nanophotonics, Optics (imaging, holography, lenses), Wireless communication (RIS, intelligent reflection surfaces), Invisibility cloaks and stealth, Sensing and detection (spectral recovery), Computational imaging and image classification, Analogue / wave-based computing and optical neural networks, Acoustics, water waves, and heat flow (cross-physical applications), Quantum mechanics / many-body physics (AI applied to physical discovery), Autonomous systems / augmented reality (low-latency, high-throughput processing) |
223. InterPLM: Discovering Interpretable Features in Protein Language Models via Sparse Autoencoders, Preprint (January 28, 2025)
| Category | Items |
|---|---|
| Datasets | UniRef50 (random sample), Swiss-Prot (reviewed subset, sampled), AlphaFold Database (structures) |
| Models | Transformer, Autoencoder, Transformer |
| Tasks | Representation Learning, Feature Extraction, Feature Selection, Clustering, Dimensionality Reduction, Binary Classification, Regression, Language Modeling, Text Generation, Clustering, Hyperparameter Optimization |
| Learning Methods | Self-Supervised Learning, Unsupervised Learning, Transfer Learning, Representation Learning, Feature Learning, Pre-training, In-Context Learning, Prompt Learning, Hyperparameter Optimization |
| Performance Highlights | features_with_strong_concept_alignment_per_layer: up to 2309, features_identified_by_SAE_vs_neurons_increase: SAEs extract 3x the concepts found in ESM-2-8M neurons and 7x the concepts found in ESM-2-650M neurons, number_of_distinct_SwissProt_concepts_detected_by_SAEs: 143 (expanded from 15 by neurons), SwissProt_concepts_evaluated: 433 concepts evaluated, example_feature_F1_high: f/1503 F1 = 0.998, example_feature_F1_medium: f/??? F1 = 0.793, 0.611 (other beta-barrel features), glycine_specific_features_F1: 0.995, 0.990, 0.86, validation_selection_threshold: feature-concept pairs with F1 > 0.5 counted per layer, steering_effect_examples: steering increases P(Glycine) at steered and nearby periodic positions; periodic glycine features propagated effect for multiple repeats with diminishing intensity, example_correlation_r_values_on_steering_fig7: r = .19, r = .16, r = .24, r = .004 (shown in figure panels for steering experiments), median_Pearson_r_on_validation_of_LLM_predictions: 0.72 (median Pearson r correlation across diverse proteins / 1200 features), example_feature_Pearson_r_values: examples: 9390 r=0.98, 10091 r=0.83, 9047 r=0.80, 4616 r=0.76, 4360 r=0.75, example_L0_values: ESM-2-8M L1 L0=128, L2=163, L3=100, L4=106, L5=134, L6=178; ESM-2-650M L1 L0=50, L9=211, L18=190, L24=121, L30=182, L33=148, example_percent_loss_recovered: values per-layer: e.g., ESM-2-8M L1 %LossRecovered=99.73, L2=99.72, L3=99.40, L4=98.94, L5=99.55, L6=100.00; 650M layer values include 99.83, 99.49, 94.28, 92.42, 96.24, 100.00 |
| Application Domains | Protein biology, Structural biology, Protein engineering / design, Computational biology / bioinformatics, Model interpretability / mechanistic interpretability |
222. Probing out-of-distribution generalization in machine learning for materials, Communications Materials (January 11, 2025)
| Category | Items |
|---|---|
| Datasets | Materials Project (MP), JARVIS, OQMD |
| Models | Random Forest, XGBoost, Graph Neural Network, Multi-Layer Perceptron, Transformer |
| Tasks | Regression, Dimensionality Reduction, Feature Extraction, Anomaly Detection |
| Learning Methods | Supervised Learning, Representation Learning, Ensemble Learning, Pre-training, Fine-Tuning, Out-of-Distribution Learning |
| Performance Highlights | MAE_MP_(eV/atom): 0.033, R2MP: 0.996, MAE_JARVIS(eV/atom): 0.036, R2JARVIS: 0.995, MAE_OQMD(eV/atom): 0.020, R2OQMD: 0.998, leave-one-element-out_R2>0.95fraction_on_MP: 85%, structure-based_leave-one-space-group_out_R2>0.95fraction: 100% (ALIGNN achieves R2 > 0.95 in all tasks; 88% of tasks have R2 > 0.98), MAE_MP(eV/atom): 0.078, R2MP: 0.979, MAE_JARVIS(eV/atom): 0.074, R2JARVIS: 0.981, MAE_OQMD(eV/atom): 0.070, R2OQMD: 0.987, leave-one-element-out_R2>0.95fraction_on_MP: 68%, MAE_MP(eV/atom): 0.090, R2MP: 0.970, MAE_JARVIS(eV/atom): 0.099, R2JARVIS: 0.968, MAE_OQMD(eV/atom): 0.065, R2OQMD: 0.985, MAE_MP(eV/atom): 0.052, R2MP: 0.992, MAE_JARVIS(eV/atom): 0.081, R2JARVIS: 0.985, MAE_OQMD(eV/atom): 0.038, R2OQMD: 0.995, MAE_MP(eV/atom): 0.063, R2MP: 0.981, MAE_JARVIS(eV/atom): 0.068, R2JARVIS: 0.982, MAE_OQMD(eV/atom): 0.045, R2_OQMD: 0.995, representationally_ID_tasks_behavior: neural scaling laws hold (ID and representationally ID OOD errors decrease with more data/compute), representationally_OOD_tasks_behavior: scaling marginally beneficial or adverse; e.g., OOD MAE increases with more training beyond ~20 epochs for some tasks, example_5x_increase: 5-fold increase in OOD MAE for leave-H-out when training set size increased from 10^4 to 10^6 (OQMD) |
| Application Domains | Materials science, Computational materials discovery, Computational chemistry / materials property prediction |
221. Integrating artificial intelligence with mechanistic epidemiological modeling: a scoping review of opportunities and challenges, Nature Communications (January 10, 2025)
| Category | Items |
|---|---|
| Datasets | COVID-19 datasets, Influenza datasets, Dengue datasets, HIV datasets, Synthetic datasets generated by epidemiological models, Social media content (self-reported symptom tweets, mobility info), Google Search Trends, Satellite imagery / remote sensing data, Electronic health records / emergency department reports, Mobility data / origin-destination matrices |
| Models | Long Short-Term Memory, Recurrent Neural Network, Graph Neural Network, Convolutional Neural Network, Multi-Layer Perceptron, Decision Tree, Support Vector Machine, Generative Adversarial Network, Graph Convolutional Network, Attention Mechanism, Random Forest, Gradient Boosting Tree, Feedforward Neural Network, Bayesian Network |
| Tasks | Infectious disease forecasting, Model parameterization and calibration, Disease intervention assessment and optimization, Retrospective epidemic course analysis, Transmission inference, Outbreak detection |
| Learning Methods | Supervised Learning, End-to-End Learning, Ensemble Learning, Reinforcement Learning, Variational Inference, Pre-training, Simulation-based inference |
| Performance Highlights | None |
| Application Domains | Infectious disease epidemiology, Public health planning and response, Epidemic forecasting and surveillance, Intervention design and optimization (vaccination, NPIs), Transmission network analysis / contact tracing, Vector-borne disease risk mapping (climate/environmental drivers), Agent-based simulation and policy evaluation |
220. Transforming the synthesis of carbon nanotubes with machine learning models and automation, Matter (January 08, 2025)
| Category | Items |
|---|---|
| Datasets | Standardized CVD experimental database (CARCO), Virtual experiments (digital twin outputs), Carbon-materials literature corpus (for model fine-tuning), Ion-implantation experimental sample pool, Characterization dataset (SEM, Raman, XPS, HRTEM, AFM) |
| Models | Transformer, GPT, BERT, Random Forest, Gradient Boosting Tree, XGBoost, Decision Tree, Generalized Linear Model |
| Tasks | Recommendation, Optimization, Regression, Binary Classification, Synthetic Data Generation, Hyperparameter Optimization |
| Learning Methods | Fine-Tuning, Pre-training, Self-Supervised Learning, Supervised Learning, Transfer Learning, Ensemble Learning, Representation Learning, Hyperparameter Optimization |
| Performance Highlights | R2: 0.67, R2: 0.65, R2: 0.64, Spearman_correlation_on_test_questions: increase from 0.1 to 0.3, Density_control_precision_workflow: 56.25% (27/49), Regression_only_precision: 39.74% (31/79), Human_filtering_precision: 49.15% (29/60), Human_after_classification_precision: 61.36% (27/45) |
| Application Domains | Carbon-based nanomaterials (CBNs), Horizontally aligned carbon nanotube (HACNT) array synthesis, Materials science and nanomaterials synthesis, Catalyst discovery and screening, Automated/robotic chemical vapor deposition (CVD) systems, Electronics and optoelectronics (applications of HACNT arrays), Biomedical sensors (application context) |
219. Synthesis Strategies for High Entropy Nanoparticles, Advanced Materials (January 08, 2025)
| Category | Items |
|---|---|
| Datasets | None |
| Models | Non-negative Matrix Factorization |
| Tasks | None |
| Learning Methods | None |
| Performance Highlights | None |
| Application Domains | Materials Science, Nanoparticle synthesis, Catalysis, Electrocatalysis, Photocatalysis, Energy storage (Batteries, Supercapacitors), Thermoelectrics, Photovoltaics, Biomedical coatings/implants, Aerospace and coatings, Nuclear reactors |
218. Development and validation of a real-time prediction model for acute kidney injury in hospitalized patients, Nature Communications (January 02, 2025)
| Category | Items |
|---|---|
| Datasets | Combined multicenter EHR cohort (derivation + internal + external), Derivation cohort (Site 1), Internal validation cohort (Site 1, 2020), External validation cohorts (Sites 2-5) |
| Models | Random Forest, LightGBM |
| Tasks | Binary Classification, Classification, Survival Analysis, Time Series Forecasting, Feature Selection, Hyperparameter Optimization |
| Learning Methods | Supervised Learning, Imbalanced Learning, Ensemble Learning, Hyperparameter Optimization |
| Performance Highlights | Derivation AUC (AKI in 24 h, test set): 0.92 (95% CI, 0.90–0.93), Derivation AUC (AKI in 48 h, test set): 0.91 (95% CI, 0.90–0.92), Derivation AUC (AKI in 72 h, test set): 0.91 (95% CI, 0.90–0.91), Derivation AUC (Severe AKI in 24 h, test set): 0.95 (95% CI, 0.94–0.97), Derivation AUC (Severe AKI in 48 h, test set): 0.95 (95% CI, 0.94–0.96), Derivation AUC (Severe AKI in 72 h, test set): 0.94 (95% CI, 0.94–0.95), Transported model AUC range (AKI within 48 h, external sites): 0.74–0.85 (per-site: Table 2 shows Site1 0.85; Site2 0.78; Site3 0.74; Site4 0.81; Site5 0.78), Transported model AUC range (Severe AKI within 48 h, external sites): 0.83–0.90 (per-site: Table 2 shows Site1 0.90; Site2 0.85; Site3 0.83; Site4 0.89; Site5 0.86), Re-fitted model AUC range (AKI within 48 h, validation cohorts): 0.81–0.90 (per-site: Table 2: Site1 0.90; Site2 0.86; Site3 0.81; Site4 0.89; Site5 0.90), Re-fitted model AUC range (Severe AKI within 48 h, validation cohorts): 0.88–0.95 (per-site: Table 2: Site1 0.95; Site2 0.92; Site3 0.88; Site4 0.93; Site5 0.91), Derivation AUC (Severe AKI in 48 h, test set): 0.95 (95% CI, 0.94–0.96), Re-fitted model AUC (Severe AKI in 48 h) Site1 (internal): 0.95 (95% CI, 0.94–0.95), Re-fitted model AUC (Severe AKI in 48 h) Site2: 0.92; Site3: 0.88; Site4: 0.93; Site5: 0.91, Probability cutoff: 0.45, Median lead-time to AKI (Site1): 72 hours (IQR 24–198), Sensitivity (Site1, cutoff 0.45): 89.4%, Specificity (Site1, cutoff 0.45): 89.3%, Negative Predictive Value (NPV, Site1, cutoff 0.45): 99.6%, Positive Predictive Value (PPV, Site1, cutoff 0.45): 24.1%, Probability cutoff: 0.4, Median lead-time to severe AKI (Site1): 114 hours, Sensitivity (Site1, cutoff 0.4): 90.9%, Specificity (Site1, cutoff 0.4): 93.9%, NPV (Site1, cutoff 0.4): 99.8%, PPV (Site1, cutoff 0.4): 19.5%, AKI within 48 h AUC (before re-re-fitting, Site3): 0.81 (0.80–0.81), AKI within 48 h AUC (after re-re-fitting, Site3): 0.89 (0.89–0.89), Severe AKI within 48 h AUC (before re-re-fitting, Site3): 0.88 (0.87–0.88), Severe AKI within 48 h AUC (after re-re-fitting, Site3): 0.90 (0.89–0.90), Specificity (AKI detection, cutoff 0.45) improved from: 66.4% to 82.6%, Sensitivity (AKI detection) improved from: 78.7% to 80.8%, PPV improved from: 19.1% to 31.9%, NPV improved from: 96.9% to 97.7% |
| Application Domains | Healthcare, Nephrology (acute kidney injury prediction), Hospital operational care / in-hospital clinical decision support, Electronic Health Records (EHR)-based predictive analytics, Clinical risk stratification and preventive medicine |
217. Machine learning for the physics of climate, Nature Reviews Physics (January 2025)
| Category | Items |
|---|---|
| Datasets | ERA5 reanalysis, Landsat-1 / satellite altimeter observations (Jason, ERS, TOPEX/Poseidon), SWOT (Surface Water and Ocean Topography) mission data, CryoSat-2 and SMOS merged product (CS2SMOS), Argo floats (including BGC-Argo), nextsim (neXtSIM) sea-ice thickness product / Arctic sea-ice forecasting, NATL60 (SWOT Data Challenge NATL60), IMERG precipitation product, High-resolution numerical simulations and process-resolving simulation libraries (e.g., large-eddy simulations library) |
| Models | Convolutional Neural Network, U-Net, Transformer, ResNet, Graph Neural Network, Recurrent Neural Network, Generative Adversarial Network, Diffusion Model, Normalizing Flow, Gaussian Process |
| Tasks | Time Series Forecasting, Image Super-Resolution, Regression, Distribution Estimation, Hyperparameter Optimization, Anomaly Detection, Causal Inference |
| Learning Methods | Supervised Learning, Online Learning, Reinforcement Learning, Transfer Learning, Variational Inference, Generative Learning, Self-Supervised Learning, Pre-training, Ensemble Learning |
| Performance Highlights | lead_time_months_for_ENSO_skill: up to 17 months (Ham et al. 2019), extended_lead_time_with_loss_and_params: up to 24 months (Patil et al. 2023), lead_time_months_reservoir_methods: reservoir computing methods reached ~21 months for ENSO (Hassanibesheli et al. 2022), lead_time_months_for_ENSO_skill: up to 18 months reported for adaptive graph CNNs (ref. 138), improvement_over_traditional_methods: outperforming traditional algorithms such as kriging (general statement; specific numeric metrics not provided), capability: can generate stochastic high-resolution samples and represent uncertainty (qualitative), examples: GAN-based stochastic super-resolution for precipitation/clouds (refs. 31,33), use_case: diffusion-based ensemble forecasting (Gencast) proposed for medium-range weather, objective: used to objectively tune parameters of parameterization schemes (qualitative benefit), bias_reduction_example: NeuralGCM shows promise in reducing some biases of traditional GCMs (qualitative), stability_issues: offline-trained CNNs can be unstable when coupled; mixed offline–online retraining produced stable QBO in testbed (ref. 81), comparison_with_IFS: global transformer S2S model outperforms IFS in key variables including total precipitation and tropical cyclones (qualitative; no numeric score provided) |
| Application Domains | Climate physics, Weather forecasting (nowcasting, medium-range), Sub-seasonal to seasonal forecasting (S2S), Interannual forecasting (ENSO prediction), Decadal forecasting, Oceanography (sea surface height, eddies, ocean turbulence), Cryosphere (sea-ice thickness), Remote sensing / satellite data reconstruction, Parameterization of sub-grid-scale processes in climate models, Data assimilation and reanalysis construction, Model emulation and hybrid physics–ML models |
215. AI4Materials: Transforming the landscape of materials science and enigneering, Review of Materials Research (January 01, 2025)
| Category | Items |
|---|---|
| Datasets | Materials Project, Open Quantum Materials Database (OQMD), NOMAD, Materials Data Curation System (MDCS), MGEDATA, Materials Cloud, MATCLOUD / MatCloud, ALKEMIE-Matter Cloud, RXN SMILES / Reaction corpora used by IBM RXN, Corpus for SteelBERT, Hypothetical polymer dataset (polyBERT training), NYU abstracts corpus (skip-gram), DeepMind / GNoME training set (first-principles calculations), Autonomous experiment logs / execution datasets |
| Models | Random Forest, Gradient Boosting Tree, Support Vector Machine, Multi-Layer Perceptron, Graph Neural Network, Convolutional Neural Network, Transformer, BERT, GPT, Variational Autoencoder, Generative Adversarial Network, Diffusion Model, Recurrent Neural Network, Bayesian methods (as Active Learning), Evolutionary Learning |
| Tasks | Regression, Classification, Clustering, Dimensionality Reduction, Feature Selection, Feature Extraction, Optimization, Experimental Design, Sequence-to-Sequence, Text Classification, Text Generation, Named Entity Recognition, Image Classification, Ranking, Data Generation |
| Learning Methods | Supervised Learning, Unsupervised Learning, Active Learning, Reinforcement Learning, Transfer Learning, Pre-training, Fine-Tuning, Multi-Task Learning, Self-Supervised Learning, Evolutionary Learning, Federated Learning |
| Performance Highlights | prediction_accuracy: 92%, experiments_per_day: approximately 100, parameter_identification_runs: 237 experiments for sensitivity stratification; >600 closed-loop experiments for growth-rate studies, screened_compositions: 19,841 compositions screened, selected_candidates: 151 FPV perovskites identified (downstream regression applied), experimental_workload_reduction: substantial (qualitative), accuracy_improvement: reported improved predictive accuracy over naive approaches, screening_time_reduction: reduced materials screening cycle by approximately ten years (qualitative), scale_of_simulation: molecular dynamics to 100 million atoms, speedup_equivalent: reduced a task that would normally take 60 years to one day (qualitative/inferred by scale), award: 2020 Gordon Bell Prize (team using deep potential approaches), capabilities: predict forward and reverse reactions and yields; rapid and accurate atomic mappings, deployment: used in RXN platform for synthesis planning, R2_on_mechanical_properties: around 80% (yield strength, tensile strength, elongation), fine-tuning_samples: as few as 64 experimental samples for austenitic stainless steels, predicted_steel_properties: yield strength 960 MPa, tensile strength 1138 MPa, elongation 32.5%, A-Lab_success_rate: 41 novel compounds synthesized out of 58 targets (~70.7%) over 17 days, ChatMOF_metrics: text-based searching 96.9%; property predicting 95.7%; structure generating 87.5% (with GPT-4), generated_candidates_reported: e.g., 200 potential high-Tc superconductors proposed; generation of eutectic compositionally complex alloys across quaternary to senary systems (quantities reported in cited works), application_examples: successful inverse design of SMAs, metamaterials, superconductors, applications: GAN used for copper-based metallic glasses, bulk metallic glass inverse design, qualitative_outcomes: enabled generation of novel amorphous solids and metallic glass compositions, improved_prediction: enhanced predictive modeling of creep fracture life (qualitative improvement reported), defect_detection_case: LLM-enabled microstructure optimization combined with ML for defects classification (paper references improved outcomes) |
| Application Domains | Materials Science and Engineering, Alloy Design and Metallurgy, Additive Manufacturing / 3D Printing, Catalysis and Photocatalysis, Batteries and Energy Storage (Li-S, solid-state conductors), Superconductors discovery, Metamaterials and Mechanical Metamaterials, Nanotechnology and Quantum Dots, Aerospace materials, Biomedical materials, High-throughput experimental automation |
214. Computational microscopy with coherent diffractive imaging and ptychography, Nature (January 2025)
| Category | Items |
|---|---|
| Datasets | Rotavirus single-particle dataset (~500 particles), Apoferritin cryo-electron ptychography dataset, Mimivirus XFEL single-particle dataset, Twisted bilayer MoS2 electron ptychography dataset, PrScO3 multislice ptychography dataset, Zeolite Socony Mobil-5 catalyst dataset (electron multislice ptychography), La3Ni2O7−δ multislice ptychography + EELS dataset, Lithium- and manganese-rich (LMR) layered-oxide in situ BCDI dataset, Nanoparticle superlattice X-ray ptychography + tomography dataset, Integrated circuit ptychographic X-ray tomography dataset (7-nm commercial IC), Fourier ptychography tissue dataset (digital pathology), Frozen–hydrated mouse brain tissue X-ray ptychographic tomography dataset |
| Models | Convolutional Neural Network, Autoencoder, Generative Adversarial Network, Multi-Layer Perceptron |
| Tasks | Image-to-Image Translation, Image Generation, Clustering, Experimental Design, 3D Reconstruction (mapped to Image-to-Image Translation) |
| Learning Methods | Representation Learning, Reinforcement Learning, Maximum Likelihood Estimation, Adversarial Training, Supervised Learning |
| Performance Highlights | real-time: claimed (contextual) |
| Application Domains | Materials science (atomic-resolution imaging, strain and defect mapping), Quantum materials and magnetism (spin textures, skyrmions, topological defects), Battery and energy materials (electrodes, in situ BCDI during cycling), Nanomaterials (nanoparticle lattices, superlattices), Integrated circuits and nanoelectronics (non-destructive 3D metrology), Structural biology (proteins, viruses, cells, cryo-electron ptychography), Biomedical imaging / Digital pathology (Fourier ptychography, tissue imaging), Ultrafast dynamics (pump–probe HCDI, XFEL single-shot experiments), Computational microscopy / inverse problems (phase retrieval and reconstruction algorithms) |
213. Probabilistic weather forecasting with machine learning, Nature (January 2025)
| Category | Items |
|---|---|
| Datasets | ERA5 reanalysis (analysis), ENS (ECMWF ensemble forecast, TIGGE archive), HRES-fc0 (ECMWF deterministic initial conditions / HRES dataset), Global Power Plant Database (GPPD), IBTrACS (International Best Track Archive for Climate Stewardship), TempestExtremes tracked cyclone outputs (from ERA5, HRES-fc0, GenCast, ENS), Derived pooled verification datasets (pooled CRPS; neighbourhood verification) |
| Models | Denoising Diffusion Probabilistic Model, Graph Neural Network, Transformer, Encoder-Decoder, Multi-Layer Perceptron, Message Passing Neural Network, Gaussian Process |
| Tasks | Time Series Forecasting, Distribution Estimation, Regression, Sequence-to-Sequence, Synthetic Data Generation, Distribution Estimation |
| Learning Methods | Generative Learning, Supervised Learning, Pre-training, Fine-Tuning, Backpropagation, Representation Learning |
| Performance Highlights | CRPS_targets_better_than_ENS_pct: 97.2%, CRPS_targets_better_than_ENS_pct_lead_gt_36h: 99.6%, ensemble_mean_RMSE_better_or_equal_pct: 96%, ensemble_mean_RMSE_significantly_better_pct: 78%, runtime_single_15-day_forecast: 8 minutes (on Cloud TPUv5), ensemble_size: 50 members, significance: P < 0.05 for reported significant comparisons, pooled_average_CRPS_better_pct: 98.1% of 5,400 pooled targets, pooled_max_CRPS_better_pct: 97.6% of 5,400 pooled targets, spectral_matching: GenCast samples spectra closely match ERA5 at 1- and 15-day lead times (qualitative; fig.2 and text), CRPS_targets_better_or_competitive_pct_vs_ENS: 82% of scorecard targets, outperformed_by_GenCast_pct: GenCast outperforms GenCast-Perturbed in 99% of targets, sample_sharpness: GenCast-Perturbed ensemble members are blurrier; ensemble-mean-like samples (qualitative), relative_CRPS_improvement_vs_ENS_up_to_2d: ~20% better, relative_CRPS_improvement_vs_ENS_2-4d: 10–20% better, statistically_significant_improvement_out_to: 7 days (P < 0.05), ensemble_mean_track_position_advantage: approx. 12-hour advantage in accuracy between 1 and 4 days ahead (GenCast ensembles mean more accurate than ENS), REV_track_probability: GenCast track probability forecasts outperform ENS (better REV at all cost/loss ratios except when neither model beats climatology); significant improvements out to 7 day lead times (P < 0.05) |
| Application Domains | Operational weather forecasting / medium-range global weather prediction, Tropical cyclone track forecasting and hazard prediction, Renewable energy forecasting (regional wind power aggregation and decision support), Meteorological verification and probabilistic forecast evaluation, Climate and atmospheric reanalysis-informed ML model training |
211. Data extraction from polymer literature using large language models, Communications Materials (December 19, 2024)
| Category | Items |
|---|---|
| Datasets | corpus of ~2.4 million materials science journal articles, subset of 681,000 polymer-related articles, filtered paragraphs after heuristic and NER filters, manually curated labeled subset for evaluation (630 abstracts), evaluation subset of 1000 polymer-related articles, extracted polymer-property dataset (Polymer Scholar) |
| Models | BERT, GPT, Transformer, Long Short-Term Memory, Recurrent Neural Network |
| Tasks | Named Entity Recognition, Information Retrieval, Text Generation, Clustering, Feature Extraction |
| Learning Methods | Few-Shot Learning, Pre-training, Fine-Tuning, Self-Supervised Learning, Supervised Learning, In-Context Learning, Prompt Learning |
| Performance Highlights | F1_Tg: 0.67, F1_bandgap_random: 0.87, F1_bandgap_similar: 0.85, extracted_records_full_corpus: 672,449, extracted_pairs_subset_6179_paragraphs_random_shot: 4706, extracted_pairs_subset_6179_paragraphs_similar_shot: 4589, full_text_Tg_records: 125,585, full_text_bandgap_records: 63,361, api_cost_for_6179_paragraphs: $4.48 (for ~2.9 million tokens, reported in Results), full_corpus_inference_cost: ≈$1,200 for 716,000 paragraphs (Methods), F1_Tg: 0.63, F1_bandgap: 0.66, extracted_records_full_corpus: 390,813, full_text_Tg_records: 75,722, full_text_bandgap_records: 30,732, processing_time_6179_paragraphs: < 30 minutes, monetary_cost: $0 (operated in-house), F1_Tg: 0.64, F1_bandgap: 0.77, extracted_pairs_subset_6179_paragraphs: 3441, inference_time: longest among evaluated (hosted locally on 4x Quadro GP100 GPUs), monetary_cost: $0 (hosted locally but high compute/time cost) |
| Application Domains | polymer science, materials science, materials informatics / polymer informatics, natural language processing (applied to scientific literature), scientific data curation / dataset creation for ML |
210. De novo design of polymer electrolytes using GPT-based and diffusion-based generative models, npj Computational Materials (December 19, 2024)
| Category | Items |
|---|---|
| Datasets | HTP-MD dataset, PI1M dataset, Generated candidates (this work), Top candidates validated with MD (this work) |
| Models | GPT, Transformer, Diffusion Model, Denoising Diffusion Probabilistic Model, BERT, Graph Neural Network, Random Forest, U-Net |
| Tasks | Language Modeling, Text Generation, Synthetic Data Generation, Regression, Binary Classification, Data Augmentation |
| Learning Methods | Self-Supervised Learning, Pre-training, Fine-Tuning, Transfer Learning, Supervised Learning, Imbalanced Learning, Hyperparameter Optimization, Multi-Objective Learning |
| Performance Highlights | unconditional_mean_metric: 0.773, validity: >0.8 (reported for optimal minGPT in unconditional generation), uniqueness: >0.8 (reported), novelty: >0.8 (reported), synthesizability: >0.8 (reported), training_time: 3-4 minutes on Tesla V100 (optimal minGPT model), pretraining_effects: shortened fine-tuning convergence; improved validity & uniqueness especially with limited fine-tuning data, unconditional_mean_metric: 0.736, validity: lower than minGPT and diffusion-LM (qualitative), training_time: ~2 hours on Tesla V100 (optimal 1Ddiffusion model), unconditional_mean_metric: 0.767, validity: >0.8 (reported for optimal diffusion-LM in unconditional generation), training_time: ~2 hours on Tesla V100 (optimal diffusion-LM model), generated_set_size: 100000, selected_for_MD: 50, MD_successful_simulations_obtained: 46, simulations_completed: 206 out of the 250 simulations across the 46 polymers (reported), candidates_surpassing_best_train_conductivity: 17, best_generated_conductivity_S/cm: 0.00113, best_train_conductivity_S/cm: 0.000507, relative_performance: GNN out-performed Random Forest (no numeric values provided), usage: used to screen 100K generated candidates and rank top 50 for MD |
| Application Domains | polymer electrolytes for lithium-ion / solid-state batteries, polymer informatics, materials discovery (computational materials), generative molecular design, high-throughput computational screening |
209. Poseidon: Efficient Foundation Models for PDEs, Advances in Neural Information Processing Systems (December 16, 2024)
| Category | Items |
|---|---|
| Datasets | Pretraining collection: NS-Sines, Pretraining collection: NS-Gauss, Pretraining collection: CE-RP (4-quadrant Riemann problem), Pretraining collection: CE-CRP (curved Riemann partitions), Pretraining collection: CE-KH (Kelvin-Helmholtz), Pretraining collection: CE-Gauss, POSEIDON pretraining aggregate (authors’ description), Downstream: NS-PwC (Piecewise-constant vorticity), Downstream: NS-BB (Brownian Bridge initializations), Downstream: NS-SL (Shear Layer), Downstream: NS-SVS (Sinusoidal Vortex Sheet), Downstream: NS-Tracer-PwC (passive tracer transport), Downstream: FNS-KF (Forced Navier-Stokes / Kolmogorov Flow), Downstream: CE-RPUI (Riemann problem with uncertain interfaces), Downstream: CE-RM (Richtmyer–Meshkov), Downstream: GCE-RT (Gravitational compressible Euler, Rayleigh–Taylor), Downstream: Wave-Gauss (wave equation in Gaussian medium), Downstream: Wave-Layer (wave equation in layered medium), Downstream: ACE (Allen–Cahn equation), Downstream: SE-AF (steady Euler flow past airfoil), Downstream: Poisson-Gauss, Downstream: Helmholtz |
| Models | Vision Transformer, Transformer, Attention Mechanism, Multi-Head Attention, U-Net, Convolutional Neural Network, Encoder-Decoder, Sequence-to-Sequence |
| Tasks | Time Series Forecasting, Sequence-to-Sequence, Image-to-Image Translation, Regression, Time Series Forecasting |
| Learning Methods | Pre-training, Fine-Tuning, Transfer Learning, Supervised Learning, Stochastic Gradient Descent, Representation Learning, Batch Learning, Mini-Batch Learning |
| Performance Highlights | median_samples_to_match_FNO_1024: 20 (median over downstream tasks, POSEIDON-L as reported), overall_best_performance_count: 14/15 tasks (POSEIDON family best on 14 of 15 tasks; POSEIDON-L top performer overall), pretraining_examples_after_all2all: approx 5.11M training examples (pretraining dataset after all2all pairing), aggregate_median_EG: 49.8 (median Efficiency Gain for POSEIDON-L over tasks), aggregate_mean_AG: 9.58 (mean Accuracy Gain for POSEIDON-L over tasks), EG: 890.6, AG: 24.7, context_metric_baseline: FNO baseline EG/AG normalized to 1 (used as reference), EG: 502.9, AG: 7.3, EG: 552.5, AG: 29.3, EG: 21.9, AG: 5.5, EG: 49.8, AG: 8.7, EG: 62.5, AG: 7.4, EG: 352.2, AG: 6.5, EG: 4.6, AG: 1.2, EG: 3.4, AG: 1.2, EG: 5.3, AG: 2.0, EG: 46.5, AG: 6.1, EG: 62.1, AG: 5.6, EG: 17.0, AG: 11.6, EG: 42.5, AG: 20.5, EG: 78.3, AG: 6.1, reference_model: FNO (baseline), normalized EG=1, AG=1 by construction (used as reference in EG/AG metrics) |
| Application Domains | Computational fluid dynamics (incompressible Navier–Stokes), Compressible gas dynamics (Euler equations), Wave propagation / acoustics / seismic (wave equation), Reaction–diffusion systems (Allen–Cahn / material science), Aerofoils / aerodynamic steady-state flow (shape optimization), Astrophysical fluid instabilities (Rayleigh–Taylor), Elliptic problems (Poisson, Helmholtz), Scientific machine learning / operator learning for PDEs |
208. Navigating Chemical Space with Latent Flows, Advances in Neural Information Processing Systems (December 16, 2024)
| Category | Items |
|---|---|
| Datasets | MOSES, ZINC250K, ChEMBL, Aggregated dataset (MOSES + ZINC250K + ChEMBL), Synthetic / sampled subsets used in experiments |
| Models | Variational Autoencoder, Multi-Layer Perceptron, Support Vector Machine, Normalizing Flow, Generative Adversarial Network, Denoising Diffusion Probabilistic Model |
| Tasks | Optimization, Regression, Classification, Data Generation, Representation Learning |
| Learning Methods | Supervised Learning, Unsupervised Learning, Pre-training, Fine-Tuning, Stochastic Gradient Descent, Evolutionary Learning, Representation Learning |
| Performance Highlights | PLOGP_top1: 4.06, PLOGP_top2: 3.69, PLOGP_top3: 3.54, QED_top1: 0.944, QED_top2: 0.941, QED_top3: 0.941, ESR1_docking_1st: -11.0, ESR1_docking_2nd: -10.67, ESR1_docking_3rd: -10.46, ACAA1_docking_1st: -9.9, ACAA1_docking_2nd: -9.64, ACAA1_docking_3rd: -9.61, PLOGP_top1: 4.76, PLOGP_top2: 3.78, PLOGP_top3: 3.71, QED_top1: 0.947, QED_top2: 0.934, QED_top3: 0.932, ESR1_docking_1st: -11.05, ESR1_docking_2nd: -10.71, ESR1_docking_3rd: -10.68, ACAA1_docking_1st: -10.48, ACAA1_docking_2nd: -10.04, ACAA1_docking_3rd: -9.88, PLOGP_top1: 5.3, PLOGP_top2: 5.22, PLOGP_top3: 5.14, QED_top1: 0.905, QED_top2: 0.902, QED_top3: 0.978, ESR1_docking_1st: -10.22, ESR1_docking_2nd: -10.06, ESR1_docking_3rd: -9.97, ACAA1_docking_1st: -9.69, ACAA1_docking_2nd: -9.64, ACAA1_docking_3rd: -9.57, PLOGP_top1: 4.39, PLOGP_top2: 3.7, PLOGP_top3: 3.48, QED_top1: 0.946, QED_top2: 0.941, QED_top3: 0.94, ESR1_docking_1st: -10.68, ESR1_docking_2nd: -10.56, ESR1_docking_3rd: -10.52, ACAA1_docking_1st: -9.89, ACAA1_docking_2nd: -9.61, ACAA1_docking_3rd: -9.6, PLOGP_top1: 4.26, PLOGP_top2: 4.1, PLOGP_top3: 4.07, QED_top1: 0.93, QED_top2: 0.928, QED_top3: 0.927, ESR1_docking_1st: -10.24, ESR1_docking_2nd: -9.96, ESR1_docking_3rd: -9.92, ACAA1_docking_1st: -9.73, ACAA1_docking_2nd: -9.31, ACAA1_docking_3rd: -9.24, PLOGP_top1: 4.74, PLOGP_top2: 3.61, PLOGP_top3: 3.55, QED_top1: 0.947, QED_top2: 0.947, QED_top3: 0.942, ESR1_docking_1st: -10.68, ESR1_docking_2nd: -10.29, ESR1_docking_3rd: -10.28, ACAA1_docking_1st: -10.34, ACAA1_docking_2nd: -9.74, ACAA1_docking_3rd: -9.64, PLOGP_top1: 3.74, PLOGP_top2: 3.69, PLOGP_top3: 3.64, QED_top1: 0.941, QED_top2: 0.936, QED_top3: 0.933, ESR1_docking_1st: -11.66, ESR1_docking_2nd: -10.52, ESR1_docking_3rd: -10.43, ACAA1_docking_1st: -9.81, ACAA1_docking_2nd: -9.72, ACAA1_docking_3rd: -9.63, PLOGP_top1: 3.52, PLOGP_top2: 3.43, PLOGP_top3: 3.37, QED_top1: 0.94, QED_top2: 0.933, QED_top3: 0.932, ESR1_docking_1st: -10.32, ESR1_docking_2nd: -10.18, ESR1_docking_3rd: -10.03, ACAA1_docking_1st: -9.86, ACAA1_docking_2nd: -9.5, ACAA1_docking_3rd: -9.34, EA(Random)_PLOGP_top1: 2.29, EA(Random)_QED_top1: 0.836, EA(ChemSpace)_PLOGP_top1: 3.79, EA(ChemSpace)_QED_top1: 0.933, EA(GradientFlow)_PLOGP_top1: 3.53, EA(GradientFlow)_QED_top1: 0.93, WAVE_UNSUP_FT_PLOGP_top1: 3.71, WAVE_UNSUP_FT_PLOGP_top2: 3.58, WAVE_UNSUP_FT_PLOGP_top3: 3.46, WAVE_UNSUP_FT_QED_top1: 0.936, WAVE_UNSUP_FT_QED_top2: 0.933, WAVE_UNSUP_FT_QED_top3: 0.933, Observation: Langevin dynamics significantly pushes entire distribution to molecules with better properties surpassing other methods in long-horizon (1000-step) optimization (Figure 3 & Figure 5)., Predictor_training_setup: trained for 20 epochs on 100k samples; validated on 10k; SGD optimizer (lr=0.001); training per predictor takes <1 minute, Usage: Surrogate predictor provides gradients used by Gradient Flow and LD; performance of traversal depends on predictor accuracy (discussed qualitatively) |
| Application Domains | Drug design / small-molecule discovery, Materials discovery (general mention), Protein-ligand binding affinity optimization (docking tasks: ESR1, ACAA1), Molecular property optimization and manipulation (chemistry / medicinal chemistry) |
206. Invariant Tokenization of Crystalline Materials for Language Model Enabled Generation, Advances in Neural Information Processing Systems (December 16, 2024)
| Category | Items |
|---|---|
| Datasets | Perov-5, Carbon-24, MP-20, MPTS-52, Combined training set (Materials Project + OQMD + NOMAD), JARVIS-DFT |
| Models | GPT, Variational Autoencoder, Diffusion Model, Normalizing Flow, Transformer |
| Tasks | Language Modeling, Text Generation, Data Generation, Synthetic Data Generation, Regression |
| Learning Methods | Self-Supervised Learning, Pre-training, Fine-Tuning, Supervised Learning, Prompt Learning |
| Performance Highlights | Perov-5one_shot_Match: 50.0%, Perov-5_one_shot_RMSE: 0.099, Carbon-24_one_shot_Match: 23.7%, Carbon-24_one_shot_RMSE: 0.169, MP20_one_shot_Match: 61.3%, MP20_one_shot_RMSE: 0.040, MPTS-52_one_shot_Match: 23.1%, MPTS-52_one_shot_RMSE: 0.109, Perov-5_20_shot_Match: 98.5%, Perov-5_20_shot_RMSE: 0.023, Carbon-24_20_shot_Match: 86.0%, Carbon-24_20_shot_RMSE: 0.148, MP20_20_shot_Match: 75.3%, MP20_20_shot_RMSE: 0.037, MPTS-52_20_shot_Match: 36.4%, MPTS-52_20_shot_RMSE: 0.088, Bandgap_condition_low_success_rate: 83.6%, Bandgap_condition_high_success_rate: 90.7%, Validity_overall_towards_low: 88.0%, Uniqueness_towards_low: 98.0%, Novelty_towards_low: 86.2%, Validity_overall_towards_high: 89.8%, Uniqueness_towards_high: 92.2%, Novelty_towards_high: 98.6%, Perov-5_one_shot_Match: 45.3%, Perov-5_one_shot_RMSE: 0.114, Carbon-24_one_shot_Match: 17.1%, Carbon-24_one_shot_RMSE: 0.297, MP20_one_shot_Match: 33.9%, MP20_one_shot_RMSE: 0.105, MPTS-52_one_shot_Match: 5.34%, MPTS-52_one_shot_RMSE: 0.211, Perov-5_20_shot_Match: 88.5%, Perov-5_20_shot_RMSE: 0.046, Carbon-24_20_shot_Match: 88.4%, Carbon-24_20_shot_RMSE: 0.229, MP20_20_shot_Match: 67.0%, MP20_20_shot_RMSE: 0.103, MPTS-52_20_shot_Match: 20.8%, MPTS-52_20_shot_RMSE: 0.209, Perov-5_one_shot_Match: 52.0%, Perov-5_one_shot_RMSE: 0.076, Carbon-24_one_shot_Match: 17.5%, Carbon-24_one_shot_RMSE: 0.276, MP20_one_shot_Match: 51.5%, MP20_one_shot_RMSE: 0.063, MPTS-52_one_shot_Match: 12.2%, MPTS-52_one_shot_RMSE: 0.179, Perov-5_20_shot_Match: 98.6%, Perov-5_20_shot_RMSE: 0.013, Carbon-24_20_shot_Match: 88.5%, Carbon-24_20_shot_RMSE: 0.219, MP20_20_shot_Match: 77.9%, MP20_20_shot_RMSE: 0.049, MPTS-52_20_shot_Match: 34.0%, MPTS-52_20_shot_RMSE: 0.175, CrystaLLM_one_shot_Perov-5_Match: 46.1%, CrystaLLM_one_shot_Perov-5_RMSE: 0.095, CrystaLLM_one_shot_Carbon-24_Match: 20.3%, CrystaLLM_one_shot_Carbon-24_RMSE: 0.176, CrystaLLM_one_shot_MP20_Match: 58.7%, CrystaLLM_one_shot_MP20_RMSE: 0.041, CrystaLLM_one_shot_MPTS-52_Match: 19.2%, CrystaLLM_one_shot_MPTS-52_RMSE: 0.111, CrystaLLM_20_shot_Perov-5_Match: 97.6%, CrystaLLM_20_shot_Perov-5_RMSE: 0.025, CrystaLLM_20_shot_Carbon-24_Match: 85.2%, CrystaLLM_20_shot_Carbon-24_RMSE: 0.151, CrystaLLM_20_shot_MP20_Match: 74.0%, CrystaLLM_20_shot_MP20_RMSE: 0.035, CrystaLLM_20_shot_MPTS-52_Match: 33.8%, CrystaLLM_20_shot_MPTS-52_RMSE: 0.106, FlowMM_validity%: 83.2%, FlowMM_stability_rate_DFT_%: 96.9%, FlowMM_S.U.N.Rate%: 2.5%, ComFormer_MAE_bandgap: 0.122 eV, MP-20_test_set_Mat2Seq_large_params: 200M, MP-20_test_set_Mat2Seq_large_RMSE: 0.037, Mat2Seq-small_params: 25M, Mat2Seq-small_RMSE: 0.039, CDVAE_params: 4.5M, CDVAE_RMSE: 0.103, DiffCSP_RMSE: 0.049, generation_speed_Mat2Seq-small_sec_per_crystal: 2.1 s, generation_speed_Mat2Seq-large_sec_per_crystal: 5.7 s, generation_speed_CDVAE_sec_per_crystal: 37.9 s, generation_speed_DiffCSP_sec_per_crystal: 7.3 s, MP-20_exp_observed_MatchRate: 65.2%, MP-20_exp_observed_RMSE: 0.042, MP-20_whole_test_set_MatchRate: 61.3%, MP-20_whole_test_set_RMSE: 0.040 |
| Application Domains | Materials discovery / computational materials science, Crystal structure prediction and generation, Band gap materials design and property-driven materials discovery, High-throughput screening of inorganic crystalline materials |
205. Crystal structure generation with autoregressive large language modeling, Nature Communications (December 06, 2024)
| Category | Items |
|---|---|
| Datasets | CrystaLLM training set (2.3M unique cell composition-space group pairs, 2,047,889 training CIF files), Challenge set (70 structures: 58 from recent literature unseen in training, 12 from training), Held-out test set (subset of the curated dataset), Perov-5 benchmark, Carbon-24 benchmark, MP-20 benchmark, MPTS-52 benchmark |
| Models | Transformer, Graph Neural Network, Variational Autoencoder, Denoising Diffusion Probabilistic Model, U-Net, Transformer |
| Tasks | Synthetic Data Generation, Data Generation, Regression |
| Learning Methods | Self-Supervised Learning, Pre-training, Supervised Learning, Fine-Tuning, Reinforcement Learning |
| Performance Highlights | held-out_test_validity_no_space_group_%: 93.8, held-out_test_validity_with_space_group_%: 94.0, space_group_consistent_no_space_group_%: 98.8, space_group_consistent_with_space_group_%: 99.1, atom_site_multiplicity_consistent_%: 99.4, bond_length_reasonableness_score_mean: 0.988, bond_lengths_reasonable_%: 94.6, average_valid_generated_length_tokens_no_sg: 331.9 ± 42.6, average_valid_generated_length_tokens_with_sg: 339.0 ± 41.4, match_with_test_structure_with_space_group_within_3attempts%: 88.1, Perov-5match_rate_n=20%CrystaLLM_a: 98.26, Perov-5_RMSE_CrystaLLM_a_n=20: 0.0236, Carbon-24_match_rate_n=20%CrystaLLM_a: 83.60, Carbon-24_RMSE_CrystaLLM_a_n=20: 0.1523, MP-20_match_rate_n=20%CrystaLLM_a: 75.14, MP-20_RMSE_CrystaLLM_a_n=20: 0.0395, MPTS-52_match_rate_n=20%CrystaLLM_a: 32.98, MPTS-52_RMSE_CrystaLLM_a_n=20: 0.1197, Perov-5_match_rate_n=20%CrystaLLM_b: 97.60, Perov-5_RMSE_CrystaLLM_b_n=20: 0.0249, Carbon-24_match_rate_n=20%CrystaLLM_b: 85.17, Carbon-24_RMSE_CrystaLLM_b_n=20: 0.1514, MP-20_match_rate_n=20%CrystaLLM_b: 73.97, MP-20_RMSE_CrystaLLM_b_n=20: 0.0349, MPTS-52_match_rate_n=20%CrystaLLM_b: 33.75, MPTS-52_RMSE_CrystaLLM_b_n=20: 0.1059, MPTS-52_match_rate_n=1_CrystaLLM_c: 28.30, MPTS-52_RMSE_n=1_CrystaLLM_c: 0.0850, MPTS-52_match_rate_n=20_CrystaLLM_c: 47.45, MPTS-52_RMSE_n=20_CrystaLLM_c: 0.0780, unconditional_generation_attempts: 1000, valid_generated_CIFs: 900, unique_structures: 891, novel_structures_vs_training_set: 102, mean_Ehull_of_102_novel_structures_eV_per_atom: 0.40, novel_structures_with_Ehull<=0.1_eV_per_atom: 20, novel_structures_with_Ehull_exact_0.00_eV_per_atom: 3, ALIGNN_used_as_predictor: formation energy per atom (used as reward in MCTS), average_ALIGNN_energy_change_after_MCTS_meV_per_atom: -153 ± 15 (prediction decrease across 102 unconditional-generated compositions), MCTS_validity_rate_improvement_no_space_group%: 95.0, MCTS_validity_rate_improvement_with_space_group_%: 60.0, MCTS_minimum_Ef_improvement_no_space_group_%: 85.0, MCTS_minimum_Ef_improvement_with_space_group_%: 65.0, MCTS_mean_Ef_improvement_no_space_group_%: 70.0, MCTS_mean_Ef_improvement_with_space_group_%: 65.0, mean_Ehull_change_after_ALIGNN-guided_MCTS_meV_per_atom: -56 ± 15 (mean Ehull improved to 0.34 eV/atom across 102 structures); 22 structures within 0.1 eV/atom of hull, successful_generation_rate_small_model_no_sg_%: 85.7, successful_generation_rate_small_model_with_sg_%: 88.6, successful_generation_rate_large_model_no_sg_%: 87.1, successful_generation_rate_large_model_with_sg_%: 91.4, match_rate_seen_small_model_%: 50.0, match_rate_seen_large_model_%: 83.3, match_rate_unseen_small_model_no_sg_%: 25.9, match_rate_unseen_small_model_with_sg_%: 34.5, match_rate_unseen_large_model_no_sg_%: 37.9, match_rate_unseen_large_model_with_sg_%: 41.4, pyrochlore_cell_parameter_R2: 0.62, pyrochlore_cell_parameter_MAE_A: 0.08 Å |
| Application Domains | Materials science, Computational materials discovery, Inorganic crystal structure prediction, Materials informatics, Computational chemistry / solid-state physics, High-throughput screening and DFT-accelerated materials design |
204. Quantifying the use and potential benefits of artificial intelligence in scientific research, Nature Human Behaviour (December 2024)
| Category | Items |
|---|---|
| Datasets | Microsoft Academic Graph (MAG), USPTO / PatentsView, Open Syllabus Project (OSP) syllabi, Survey of Doctorate Recipients (SDR) |
| Models | Convolutional Neural Network, Recurrent Neural Network, Generative Adversarial Network, Random Forest, Support Vector Machine, Decision Tree, Bayesian Network |
| Tasks | Feature Extraction, Feature Extraction, Feature Selection, Regression, Object Detection, Image Segmentation, Pose Estimation, Image Generation |
| Learning Methods | Reinforcement Learning, Fine-Tuning |
| Performance Highlights | CS_direct_AI_use_2000: 0.5%, CS_direct_AI_use_2019: 1.3%, trend_slope_b: 0.00031, trend_P: <0.001, trend_95%_CI: (0.00025, 0.00037), hit_rate_ratio_mean: 1.816, hit_rate_ratio_s.e.: 0.138, hit_rate_ratio_95%_CI: (1.547, 2.086), outside_field_citation_ratio_mean: 1.069, outside_field_citation_ratio_s.e.: 0.028, outside_field_citation_ratio_95%_CI: (1.015, 1.124), direct_vs_potential_percentile_correlation_r: 0.891, direct_vs_potential_percentile_correlation_P: <0.001, direct_vs_potential_percentile_95%_CI: (0.865, 0.913), corr_directAI_collab_r: 0.841, corr_directAI_collab_P: <0.001, corr_directAI_collab_95%_CI: (0.616, 0.939), corr_potentialAI_collab_r: 0.802, corr_potentialAI_collab_P: <0.001, corr_potentialAI_collab_95%_CI: (0.535, 0.923), example_trend_engineering_share_1990: 0.21, example_trend_engineering_share_2019: 0.44, engineering_trend_slope_b: 0.0057, engineering_trend_P: <0.001, engineering_trend_95%_CI: (0.0047, 0.0068), women_vs_directAI_r: -0.555, women_vs_directAI_P: 0.032, women_vs_directAI_95%_CI: (-0.831, -0.059), women_vs_potentialAI_r: -0.593, women_vs_potentialAI_P: 0.020, URM_vs_directAI_r: -0.734, URM_vs_directAI_P: 0.002, URM_vs_directAI_95%_CI: (-0.906, -0.355), URM_vs_potentialAI_r: -0.711, URM_vs_potentialAI_P: 0.003, example_black_vs_white_directAI_ratio_drop: Black score is 78% less than white for direct AI use (paper statement), example_black_vs_white_potentialAI_ratio_drop: Black score is 86% less than white for potential AI benefits (paper statement), hit_rate_ratio_mean: 1.816, hit_rate_ratio_s.e.: 0.138, hit_rate_ratio_95%_CI: (1.547, 2.086), outside_field_citation_ratio_mean: 1.069, outside_field_citation_ratio_s.e.: 0.028, outside_field_citation_ratio_95%_CI: (1.015, 1.124) |
| Application Domains | Computer science, Biology, Physics, Economics, Engineering, Medicine, Materials science, Mathematics, Sociology, Psychology, Political science, Geography, Geology, Chemistry, Business, Environmental science, Philosophy, History, Art |
203. Multifunctional high-entropy materials, Nature Reviews Materials (December 2024)
| Category | Items |
|---|---|
| Datasets | HEM literature corpus (natural language analysis), Abstracts corpus used in Pei et al., Candidate composition space (Pei et al.), Closed-loop active-learning experimental set (Rao et al.), High-throughput electronic calculations and experimental structure data (generic) |
| Models | Transformer, Graph Neural Network, Multi-Layer Perceptron, Gaussian Process |
| Tasks | Information Retrieval, Text Classification, Optimization, Experimental Design, Regression, Feature Extraction, Ranking |
| Learning Methods | Active Learning, Pre-training, In-Context Learning, Supervised Learning |
| Performance Highlights | abstracts_analyzed: 6.4 million, promising_compositions_identified: nearly 500 out of 2.6 million candidate compositions, new_alloys_processed: 17, discovered_property: Invar HEMs with extremely low thermal expansion coefficients (~2×10^-6 K^-1 at 300 K) |
| Application Domains | Materials science / alloy design, Magnetic materials and hard/soft magnets, Thermoelectrics, Electrocatalysis and heterogeneous catalysis, Photovoltaics / optoelectronics, Hydrogen storage and hydrides, Radiation-resistant materials, Shape-memory and multicaloric materials, Biomedical implant materials, High-throughput computational materials discovery |
202. Learning spatiotemporal dynamics with a pretrained generative model, Nature Machine Intelligence (December 2024)
| Category | Items |
|---|---|
| Datasets | Kuramoto–Sivashinsky equation (KSE) simulation dataset, Kolmogorov turbulent flow simulation dataset, ERA5 reanalysis subset (u,v,Temp,P), Cylinder flow PIV experimental dataset, Additional PDE datasets (Burgers’, Korteweg–de Vries, compressible Navier–Stokes) |
| Models | Diffusion Model, Denoising Diffusion Probabilistic Model, U-Net, Convolutional Neural Network, Graph Neural Network |
| Tasks | Image Super-Resolution, Image-to-Image Translation, Time Series Forecasting, Sequence-to-Sequence, Regression, Distribution Estimation |
| Learning Methods | Self-Supervised Learning, Pre-training, Zero-Shot Learning, Unsupervised Learning |
| Performance Highlights | nRMSE: S3GM attains lower nRMSE than baselines (FNO, U-Net, LNO, DeepONets) across downsampling factors (1× to 64×) as shown in Fig.2c, uncertainty: s.d. fields computed from five different predictions; uncertainty reported qualitatively (uncertainty fields shown), nRMSE: S3GM shows low relative errors and uncertainty when reconstructing from spectral measurements; outperforms end-to-end baselines trained for spectral-to-physical mapping (Fig.2e,f), nRMSE: S3GM produces more accurate and stable long-term predictions than baselines (FNO, U-Net); error accumulation lower over long horizons (see Fig.2i and Fig.3f), spectral_consistency: Kinetic energy spectrum of S3GM matches reference better than baselines (Fig.3g), nRMSE: nRMSE decreases as the number of observed points increases (Fig.4d) — S3GM yields acceptable nRMSE even with 1% observations plus 10% Gaussian noise, Pearson_correlation: Correlation between predictions and ground truth increases toward ~1 as observed portion increases (Fig.4e), nRMSE: S3GM statistically achieves lower nRMSE than PINN across Reynolds numbers (109.3,159.0,248.5) and as number of sensor positions varies (Fig.5d), spectral_match: Fourier features and vortex shedding frequency recovered accurately (Fig.5h), nRMSE: Baselines (including U-Net) often have higher nRMSE than S3GM across many experiments (Figs.2c,3c,3f) |
| Application Domains | fluid dynamics (turbulence, Kolmogorov flow, Navier–Stokes), spatiotemporal chaotic systems (Kuramoto–Sivashinsky equation, KdV, Burgers’), climate and atmospheric science (ERA5 reanalysis), experimental fluid mechanics / laboratory PIV measurements, general scientific and engineering full-field reconstruction from sparse sensors |
201. Towards the holistic design of alloys with large language models, Nature Reviews Materials (December 2024)
| Category | Items |
|---|---|
| Datasets | corpus of six million texts (literature mining), scientific corpora (publications, patents, conference abstracts and other corpora), structured data sets generated from automated laboratories, ImageNet (cited as an analogy / example), Protein Data Bank (cited as an analogy / example) |
| Models | Transformer, BERT, GPT, Convolutional Neural Network |
| Tasks | Named Entity Recognition, Regression, Recommendation, Optimization, Text Summarization, Information Retrieval, Ranking |
| Learning Methods | Fine-Tuning, Pre-training, Prompt Learning, Domain Adaptation, Unsupervised Learning, Representation Learning, Supervised Learning |
| Performance Highlights | gpu_hours: 1,700,000, CO2_equivalent_tons: 291 |
| Application Domains | Alloy design, Metallurgy / metallic materials, Materials science, Additive manufacturing, Sustainability assessment for materials, Materials mining and literature-based discovery |
199. An automatic end-to-end chemical synthesis development platform powered by large language models, Nature Communications (November 23, 2024)
| Category | Items |
|---|---|
| Datasets | Semantic Scholar academic literature database, High-throughput screening (HTS) experimental dataset (this work), Kinetics time-course datasets (this work), Reaction optimization experimental dataset (this work), Repository of code and processed data |
| Models | GPT, Transformer, Gaussian Process |
| Tasks | Information Retrieval, Optimization, Regression, Time Series Forecasting, Feature Extraction, Information Retrieval, Planning, Control, Optimization |
| Learning Methods | In-Context Learning, Multi-Agent Learning, Model-Based Learning, Pre-training |
| Performance Highlights | agreement_with_manual_analysis: nearly consistent, highest_identified_yield: 94.5%, PI_stopping_met_after_experiments: 36, photoredox_optimum_yield: 87%, DMSO_R2: 0.996, MeCN_R2: 0.994, DMSO_k1: 22.34, DMSO_k2: 2.84e-3, DMSO_k3: 2.51e-4, MeCN_k1: 16.29, MeCN_k2: 6.0e-3, MeCN_k3: 5.30e-4, SNAr_model: r15 = 0.2 [13][14]^2, R2: 0.995, scale_up_isolated_yield: 86%, scale_up_purity: >98%, ResultInterpreter_stop_suggestion_experiment_number: 26 (heuristic), PI_stop_experiment_number: 36 (statistical) |
| Application Domains | Chemical synthesis / synthetic chemistry, Organic chemistry (method development), Medicinal chemistry / drug discovery (reaction types relevant to drug discovery: SNAr, cross-coupling), Catalyst development, Process development and scale-up, Photoelectrochemistry / heterogeneous photoelectrocatalysis, Automation and laboratory robotics (automated HTS, OT-2, Unchained Big Kahuna) |
198. Sequence modeling and design from molecular to genome scale with Evo, Science (November 15, 2024)
| Category | Items |
|---|---|
| Datasets | OpenGenome (GTDB + IMG/VR + IMG/PR compilation), CRISPR-Cas fine-tuning dataset, IS200/IS605 fine-tuning dataset, Deep Mutational Scanning (DMS) datasets — prokaryotic protein DMS, Deep Mutational Scanning (DMS) datasets — human proteins, ncRNA DMS datasets, Promoter / RBS expression datasets (supervised and evaluation), Gene essentiality studies (DEG + phage screens), Genome-scale generation / evaluation set |
| Models | Transformer, Attention Mechanism, Multi-Head Attention, Convolutional Neural Network, Linear Model, Self-Attention Network |
| Tasks | Language Modeling, Regression, Binary Classification, Synthetic Data Generation, Feature Extraction, Data Generation, Clustering |
| Learning Methods | Self-Supervised Learning, Pre-training, Fine-Tuning, Zero-Shot Learning, Supervised Learning, Transfer Learning |
| Performance Highlights | evaluation_perplexity_range_for_baselines: eval PPL ~3.2 - 3.8 (reported across architectures in scaling laws analysis), CNN_on_one-hot_mean_Spearman_r: 0.44, CNN_on_Evo_embeddings_mean_Spearman_r: 0.56, Ridge_on_Evo_embeddings_mean_Spearman_r: not explicitly numeric in text (reported as lower than CNN); zero-shot Evo likelihood mean Spearman r = 0.43 for promoters, relative_performance: Evo’s zero-shot performance exceeded other nucleotide models and was competitive with leading protein-specific language models (per-figure; exact per-dataset Spearman values in supplemental tables), example_Spearman_r_on_5S_rRNA: Spearman r = 0.60 (two-sided t-distributed P = 1.9 × 10^-3), promoter_zero_shot_mean_Spearman_r: 0.43, promoter_GC_content_mean_Spearman_r: 0.35, GenSLM_zero_shot_mean_Spearman_r_on_promoters: 0.09, protein_expression_promoter-RBS_zero_shot_Spearman_r: 0.61, RBS_Calculator_performance_Spearman_r: 0.39, selected_candidates_tested: 11 Evo-generated Cas9 systems selected for validation; 1 generation (EvoCas9-1) exhibited robust in vitro cleavage activity comparable to SpCas9, EvoCas9-1_sequence_identity_to_closest_Cas_database: 79.9%, EvoCas9-1_sequence_identity_to_SpCas9: 73.1%, Evo_generated_sgRNA_identity_to_SpCas9_sgRNA: 91.1%, AlphaFold3_cofold_mean_pLDDT: 90.01 (EvoCas9-1 cofold), tested_designs: 24 IS200-like and 24 IS605-like designs experimentally tested, successful_IS200_like: 11 out of 24 (≈45.8%) demonstrated excision and insertion in vitro, successful_IS605_like: 3 out of 24 (12.5%) demonstrated excision and insertion in vitro, sequence_identity_minimum_successful_examples: functional designs encoded proteins with sequence identity as low as 67% to fine-tuning database, significant_association_with_essentiality: Evo log-likelihood changes with 66k context significantly associated (Bonferroni-corrected P < 0.05) with gene essentiality in 49 of 58 genomes tested, AUROC_lambda_phage: 0.90 (P < 1×10^-5), AUROC_Pseudomonas_aeruginosa: 0.84 (P < 1×10^-5), generated_sequences_count_and_size: 16 sequences ~1 Mb each (total ~16 Mb), tRNA_count_generated: 128 tRNA sequences encoding anticodons for all canonical amino acids across generated sequences, coding_density_comparison: Generated sequences have coding densities nearly the same as natural genomes and substantially higher than random sequences |
| Application Domains | Genomics, Molecular biology, Protein engineering, Synthetic biology / genome design, Metagenomics and sequence mining, Biotechnology (CRISPR and transposon tool design), Functional genomics (gene essentiality prediction) |
197. Learning the language of DNA, Science (November 15, 2024)
| Category | Items |
|---|---|
| Datasets | 2.7 million evolutionarily diverse prokaryotic and phage genomes (300 billion nucleotides), Experimental validation set: 11 Evo-generated DNA sequences |
| Models | Transformer, Attention Mechanism |
| Tasks | Language Modeling, Text Generation, Synthetic Data Generation, Regression, Classification, Sequence-to-Sequence |
| Learning Methods | Pre-training, Self-Supervised Learning, Prompt Learning, Generative Learning, Representation Learning |
| Performance Highlights | perplexity: improved (no numeric value reported); improvement of Evo over StripedHyena alone reported; perplexity improves with increasing context size, functional_validation: 1 new Cas9 (Evo-Cas9-1) functionally validated in vitro among sequences generated; 11 generated sequences were selected for experimental validation, predictive_ability: qualitative statement: ‘can predict critical features of DNA sequence, including the effect of mutations’ (no numeric metrics reported) |
| Application Domains | Genomics (prokaryotic and phage genomes), Synthetic biology, CRISPR-Cas design and protein-RNA complex design, DNA/RNA/protein function prediction, Genome-scale sequence generation and design |
196. Deep learning generative model for crystal structure prediction, npj Computational Materials (November 12, 2024)
| Category | Items |
|---|---|
| Datasets | MP60-CALYPSO, MP60 (selected from Materials Project), CALYPSO (selected), MP20, MP experimental test subset (ICSD labeled), MP experimental subset with <20 atoms, Randomly generated structures (CALYPSO) used for comparison |
| Models | Variational Autoencoder, Diffusion Model, Graph Neural Network, Message Passing Neural Network, Multi-Layer Perceptron, Variational Autoencoder (CDVAE baseline) |
| Tasks | Synthetic Data Generation, Data Generation, Optimization, Representation Learning |
| Learning Methods | Unsupervised Learning, Representation Learning, Batch Learning |
| Performance Highlights | reconstruction_match_rate_on_MP60-CALYPSO_test (%): 16.58, reconstruction_N-RMSD_on_MP60-CALYPSO: 0.2066, success_rate_on_3547MP_experimental_structures_within_800_samplings (%): 59.3, success_rate_on_2062_structures_with<20atoms_within_800_samplings (%): 83.2, reconstruction_match_rate_on_MP60-CALYPSO_test (%): 13.01, reconstruction_N-RMSD_on_MP60-CALYPSO: 0.2093, success_rate_on_3547_MP_experimental_structures_within_800_samplings (%): 46.4 (plateau approx.), success_rate_on_2062_structures_with<20_atoms_within_800_samplings (%): 69.9, DFT_local_optimization_convergence_rate_at_0GPa (%): 94.60, DFT_local_optimization_convergence_rate_at_100GPa (%): 97.00, average_ionic_steps_at_0GPa (#): 44.73, average_RMSD_after_relaxation_at_0GPa (Å): 0.79, reconstruction_match_rate_on_MP20_test (%): 45.43, reconstruction_N-RMSD_on_MP20: 0.0356, Li_cI16 (16 atoms, 50 GPa) Nmodel (avg samplings to ground state): 566.0, Li_cI16 Runs_successful_Nmodel/runs: 2/5, Li_cI16 CALYPSO_NCSP (avg samplings): 50.0 (3/3), B_alpha-B12 (36 atoms, 0 GPa) Nmodel: 74.0 (1/5 successful runs), B_gamma-B28 (28 atoms, 50 GPa) Nmodel: 341.0 (5/5 successful runs); CALYPSO: 0/3 success within 1000 samplings, SiO2_alpha-quartz (9 atoms, 0 GPa) Nmodel: 62.8 (5/5) vs CALYPSO 189.0 (3/3), SiO2_coesite (24 atoms, 5 GPa) Nmodel: 328.0 (3/5) vs CALYPSO failed within 1000 samplings |
| Application Domains | Computational materials science, Crystal structure prediction (CSP), High-pressure materials discovery, Materials design and discovery (superconductors, superhard materials), Atomistic structure generation and sampling |
195. Crystal Structure Determination from Powder Diffraction Patterns with Generative Machine Learning, Journal of the American Chemical Society (November 06, 2024)
| Category | Items |
|---|---|
| Datasets | mp-20 (Materials Project subset), RRUFF database, American Mineralogist Crystal Structure Database, Powder Diffraction File (PDF) database, High-pressure experimental PXRD data (authors’ lab / APS HPCAT beamline) |
| Models | Denoising Diffusion Probabilistic Model, Variational Autoencoder, Convolutional Neural Network, Multi-Layer Perceptron, Graph Neural Network |
| Tasks | Structured Prediction, Regression, Feature Extraction, Synthetic Data Generation |
| Learning Methods | Generative Learning, Denoising Diffusion Probabilistic Model, Variational Inference, Supervised Learning, Data Augmentation, Gradient Descent |
| Performance Highlights | simulated_match_rate_1_attempt: 30.4%, simulated_match_rate_32_attempts: 61.8%, simulated_match_rate_64_attempts: 66.6% (paper also reports up to 67% elsewhere), experimental_RRUFF_match_rate_64_attempts_with_augmentation: 41.8%, experimental_RRUFF_match_rate_64_attempts_without_augmentation: 17.9%, experimental_RRUFF_match_rate_1_attempt_with_augmentation: 8.2%, experimental_RRUFF_match_rate_1_attempt_without_augmentation: 2.2%, average_best_RMSD_over_64_attempts: 0.0397, implicit: VAE used as encoder/latent sampler in the overall model; performance metrics aggregated under Crystalyze diffusion results (see diffusion metrics), post-StructSnap_lattice_tolerance_match_rate_1%: up to 22.3%, post-StructSnap_angle_tolerance_0.5deg_match_rate: up to 26.7%, see_overall_diffusion_performance: metrics reported under primary diffusion model (e.g., up to 66.6% simulated match rate; 41.8% experimental with augmentation) |
| Application Domains | Materials science, Solid-state chemistry / crystallography, Powder X-ray diffraction (PXRD) analysis, High-pressure materials synthesis and discovery (diamond anvil cell experiments), Automated / high-throughput materials discovery |
193. Physics-Informed Inverse Design of Programmable Metasurfaces, Advanced Science (November 06, 2024)
| Category | Items |
|---|---|
| Datasets | Simulated LC metasurface designs (On-state and Off-state reflections with retrieved mode parameters), Experimental measurement data from fabricated sample (112 x 112 units) |
| Models | Multi-Layer Perceptron, Feedforward Neural Network, Autoencoder, Variational Autoencoder, Generative Adversarial Network, ResNet |
| Tasks | Regression, Optimization |
| Learning Methods | Supervised Learning, Gradient Descent, Backpropagation |
| Performance Highlights | training_dataset_size: 12000, validation_dataset_size: 3000, convergence_speed_vs_conventional_MLP: 2.5x faster, hyperparameter_scale_vs_conventional_MLP: 10x larger hyperparameters, MSE: statistically much smaller MSE distribution when physics layers are integrated (no absolute numeric provided), inverse_design_candidates_generated: 500 candidates, inverse_design_runtime: 10 s to generate 500 candidates, simulated_phase_tuning: ≈300°, simulated_reflection_amplitude_at_465GHz: >0.9 (over 90%), experimental_measured_phase_at_intersection: 264.8° (measured), experimental_measured_max_phase_change: 253.0° at 465.4 GHz (measured under 10.5° incidence), experimental_reflection_amplitude_at_intersection: ≈0.7 (measured at 467 GHz), beam_steering_max_deflection: 68° at 442 GHz (measured), 2-bit_deflection: ≈31.6° at 457 GHz with amplitude 0.26 (measured), tri-state_deflection: 23° at 457 GHz (measured) |
| Application Domains | Terahertz programmable metasurfaces / metamaterials, Beam steering for telecommunications (including 6G wireless communications), Hyperspectral imaging, Holographic displaying / holography, Photonic device design / metasurface inverse design |
192. Reproducibility in automated chemistry laboratories using computer science abstractions, Nature Synthesis (November 2024)
| Category | Items |
|---|---|
| Datasets | None |
| Models | Transformer, GPT |
| Tasks | Machine Translation, Optimization, Experimental Design, Decision Making, Control, Experimental Design |
| Learning Methods | Reinforcement Learning, Active Learning, Transfer Learning, Fine-Tuning |
| Performance Highlights | yield_labA: 87%, yield_labB: 47% |
| Application Domains | Chemistry, Materials Science, Automated Laboratory / Laboratory Robotics, Molecular Discovery, Photochemistry / Quantum Dots, Catalysis, Flow Chemistry and Batch Chemistry workflows, Analytical Chemistry (HPLC, chromatography, NMR) |
191. Autonomous mobile robots for exploratory synthetic chemistry, Nature (November 2024)
| Category | Items |
|---|---|
| Datasets | Parallel divergent synthesis screening and diversification reactions (thio)ureas, Supramolecular host–guest discovery reaction library, Photocatalyst screening for decarboxylative conjugate addition, Autonomous analytical measurement database (UPLC–MS and 1H NMR spectra) |
| Models | None |
| Tasks | Binary Classification, Feature Extraction, Novelty Detection |
| Learning Methods | None |
| Performance Highlights | None |
| Application Domains | Synthetic organic chemistry, Medicinal chemistry / Parallel diversification, Supramolecular chemistry (metal–organic assemblies, host–guest binding), Photochemical synthesis / Photocatalysis, Autonomous laboratories / Mobile robotics integration |
189. Transforming science labs into automated factories of discovery, Science Robotics (October 23, 2024)
| Category | Items |
|---|---|
| Datasets | historical and online data, massive quantities of experimental data (generated by automated labs), experimental runs / datasets produced by autonomous systems cited (e.g., AlphaFlow, mobile robotic chemist) |
| Models | Transformer, Multi-Layer Perceptron |
| Tasks | Experimental Design, Optimization, Decision Making, Policy Learning, Control, Planning, Regression, Experimental Design |
| Learning Methods | Reinforcement Learning, Supervised Learning, Prompt Learning, Representation Learning |
| Performance Highlights | None |
| Application Domains | chemistry, biochemistry, materials science, energy, catalysis, biotechnology, sustainability, electronics, drug design, semiconductor materials, batteries, photocatalysis, organic light-emitting devices (OLEDs) |
188. Open Materials 2024 (OMat24) Inorganic Materials Dataset and Models, Preprint (October 16, 2024)
| Category | Items |
|---|---|
| Datasets | OMat24 (Open Materials 2024), MPtrj (Materials Project trajectories), Alexandria, sAlexandria (subset of Alexandria), OC20 / OC22 (referenced), WBM dataset (used by Matbench-Discovery), Matbench-Discovery (benchmark) |
| Models | Graph Neural Network, Transformer |
| Tasks | Regression, Binary Classification, Classification |
| Learning Methods | Pre-training, Fine-Tuning, Transfer Learning, Supervised Learning, Self-Supervised Learning |
| Performance Highlights | F1: 0.916, MAE_energy: 0.020 eV/atom (20 meV/atom), RMSE: 0.072 eV/atom (72 meV/atom), Accuracy: 0.974, Precision: 0.923, Recall: 0.91, R2: 0.848, F1: 0.823, MAE_energy: 0.035 eV/atom (35 meV/atom), RMSE: 0.082 eV/atom (82 meV/atom), Accuracy: 0.944, Precision: 0.792, Recall: 0.856, R2: 0.802, Energy_MAE_validation: 9.6 meV/atom, Forces_MAE_validation: 43.1 meV/Å, Stress_MAE_validation: 2.3 (units consistent with meV/Å^3), Test_splits_energy_MAE_range: ≈9.7 - 14.6 meV/atom depending on test split (ID/OOD/WBM), F1: 0.86, MAE_energy: 0.029 eV/atom (29 meV/atom), RMSE: 0.078 eV/atom (78 meV/atom), Accuracy: 0.957, Precision: 0.862, Recall: 0.858, R2: 0.823, Validation_energy_MAE_on_MPtrj: 10.58 - 12.4 meV/atom depending on model variant; (Table 9: eqV2-L-DeNS energy 10.58 meV/atom; eqV2-S 12.4 meV/atom), Validation_forces_MAE_on_MPtrj: ≈30 - 32 meV/Å |
| Application Domains | Materials discovery, Inorganic materials / solid-state materials, DFT surrogate modeling (predicting energies, forces, stress), Computational screening for stable materials (thermodynamic stability / formation energy prediction), Catalyst discovery and related atomistic simulations, Molecular dynamics / non-equilibrium structure modeling (potential downstream application) |
187. MatGPT: A Vane of Materials Informatics from Past, Present, to Future, Advanced Materials (October 09, 2024)
| Category | Items |
|---|---|
| Datasets | Inorganic Crystal Structure Database (ICSD), Materials Project, AFLOW, OQMD, NOMAD, C2DB, JARVIS, ESP, OpenKIM, OMDB, ChEMBL, ZINC, GDB, Dataset for HER NN model (first-principles adsorption configurations), Dataset of 6,531 2D materials, Adsorption energies at 198 active sites, CO2 reduction / Cu–Al dataset (DFT + active learning example), Liquid electrolyte dataset for Coulomb efficiency, Garnet candidate dataset, Mechanical property dataset from MP, Perovskite synthesisability dataset, MOF pretraining set, Elpasolite compositions dataset, ZeoGAN / zeolite dataset, HEA dataset (high-entropy alloys), HMFP / IonML high-fidelity Li+ conductor dataset, Adsorption dataset for HMIs on g-C3N4 (transfer learning example) |
| Models | Decision Tree, Support Vector Machine, Random Forest, XGBoost, K-Means, Agglomerative Hierarchical Clustering, Spectral Clustering, Multi-Layer Perceptron, Convolutional Neural Network, Recurrent Neural Network, Long Short-Term Memory, Autoencoder, Deep Belief Network, Generative Adversarial Network, Variational Autoencoder, Transformer, Deep Reinforcement Learning, Gaussian Process, Graph Neural Network, Graph Convolutional Network, Message Passing Neural Network, Gradient Boosting Tree, Deep Convolutional GAN |
| Tasks | Regression, Classification, Clustering, Dimensionality Reduction, Feature Extraction, Clustering, Hyperparameter Optimization, Graph Generation, Sequence-to-Sequence, Image Generation, Optimization, Regression, Classification |
| Learning Methods | Supervised Learning, Unsupervised Learning, Semi-Supervised Learning, Contrastive Learning, Transfer Learning, Active Learning, Data Augmentation, Ensemble Learning, Pre-training, Fine-Tuning, Reinforcement Learning, Self-Supervised Learning |
| Performance Highlights | accuracy: 95.2%, out_of_sample_true_positive_rate: 0.957, RMSE_initial_discharge_capacity: 16.66 mAh/g, RMSE_50th_cycle_discharge_capacity: 18.59 mAh/g, RMSE: < 0.1 eV, selected_candidates: 38 out of 6531 2D materials screened, prediction_performance: high prediction accuracy reported for PV parameters and ablation loss (quantitative numbers not specified), state_of_the_art: claimed state-of-the-art results for a variety of properties (no single numeric metric reported), generated_structures_count: e.g., ZeoGAN generated 121 crystalline porous materials; GAN for Elpasolites and Mg–Mn–O predicted multiple novel candidate structures, success_rate_first_principles_verification: between 7.1% and 38.9% (for FTCP framework cases), classification_accuracy_double_perovskites: 92%, general_success: reported good predictive performance across many applications (specific metrics vary per study), speedup: improved the efficiency of constructing macromolecular force fields by >10^4 times (order-of-magnitude claims); ITLFF improved force-field construction efficiency by >10^4 |
| Application Domains | materials science (general), catalysis / heterogeneous catalysis, battery materials / electrolytes / solid-state electrolytes / electrode materials, photovoltaic materials / perovskites, environmental materials (adsorbents, membranes for pollutant removal), high-entropy alloys and alloy design, 2D materials, metal–organic frameworks (MOFs), organic macromolecular materials (proteins, enzymes), crystal structure prediction, autonomous experimentation / experimental robots / high-throughput synthesis |
186. Machine learning for data-centric epidemic forecasting, Nature Machine Intelligence (October 2024)
| Category | Items |
|---|---|
| Datasets | United States COVID-19 Forecast Hub dataset, Google COVID-19 Community Mobility Reports, Facebook-sampled online symptomatic surveys, Wastewater SARS-CoV-2 RNA measurements, Remote-sensing satellite imagery (hospital parking lot vacancies), Pharmaceutical and retail/supermarket sales records, CDC FluSight challenge datasets (seasonal influenza surveillance data), Reinhart et al. open repository of real-time COVID-19 indicators, Genomics / pathogen lineage datasets (phylodynamics) |
| Models | ARIMA Model, Gaussian Process, Recurrent Neural Network, Transformer, Long Short-Term Memory, Variational Autoencoder, Convolutional Neural Network, Graph Neural Network, Latent Dirichlet Allocation |
| Tasks | Time Series Forecasting, Regression, Density Estimation, Distribution Estimation, Anomaly Detection, Decision Making, Resource Allocation, Clustering |
| Learning Methods | Supervised Learning, Transfer Learning, Meta-Learning, Multi-Task Learning, Ensemble Learning, Variational Inference, Representation Learning |
| Performance Highlights | evaluation_metrics_mentioned: log score; interval score; weighted interval score; mean absolute error, qualitative: Ensembles consistently outperform individual models in multiple CDC forecasting competitions (influenza, Ebola, COVID-19); adaptive and equally weighted ensembles cited as effective |
| Application Domains | Epidemic forecasting / epidemiology, Public health decision-making, Pandemic preparedness, Healthcare resource allocation (e.g., ventilators, hospital capacity), Policy evaluation (lockdowns, travel restrictions, vaccination strategies), Supply chain planning for medical supplies |
185. Generative deep learning for the inverse design of materials, Preprint (September 27, 2024)
| Category | Items |
|---|---|
| Datasets | Open Quantum Materials Database (OQMD), Computational 2D Materials Database (C2DB) / Computational 2D Materials Database, Inorganic Crystal Structure Database (ICSD), NFFA-EUROPE SEM Dataset, ASM Micrograph Database, UHCSDB (UHCSDB / UHCSDB steel SEM dataset), DoITPoMS micrograph collection, Synthetic microstructure images by GRF (Gaussian Random Field), Ferrite-martensite SEM dataset, HMX SEM image (single large image), U-10Mo SEM-BSE dataset, Dual-phase steel synthetic micrographs, AZ80 magnesium alloy components dataset, SAOED composites micro-CT images, Large DDPM microstructure training set (Düreth et al.), 2D database of structural features and 3D porous material database (Lyu et al.), SOFC anode PFIB-SEM dataset (Hsu et al.), Li-ion battery cathode XCT and SOFC anode XCT (Gayon-Lombardo et al.), Crystal datasets used in various crystal generative works (e.g., V-O binary, Heusler, chalcogenides) |
| Models | Variational Autoencoder, Generative Adversarial Network, Denoising Diffusion Probabilistic Model, Convolutional Neural Network, Graph Neural Network, Crystal Graph Convolutional Neural Networks, U-Net, Random Forest, Support Vector Machine, Gaussian Process, Transformer, SE(3)-equivariant GNN (GemNet-dT), Diffusion Model (general) |
| Tasks | Image Generation, Image-to-Image Translation, Synthetic Data Generation, Regression, Classification, Data Generation, Graph Generation, Image Denoising, Hyperparameter Optimization, Image Super-Resolution, Structured Prediction |
| Learning Methods | Unsupervised Learning, Supervised Learning, Active Learning, Representation Learning, Fine-Tuning, Self-Supervised Learning |
| Performance Highlights | generation_success_rate: less than 1%, generation_efficiency_improvement: 100x (when adding property constraints in loss) |
| Application Domains | Materials science, Crystal structure inverse design, Microstructure generation and design, Porous materials design, Metal alloys (e.g., high-entropy alloys, NiTi shape memory alloys), Battery electrode microstructures (Li-ion cathode), Solid Oxide Fuel Cell (SOFC) anode microstructures, Metal additive manufacturing and process-structure optimization, Mechanoluminescent composites, MOF (metal-organic frameworks) / nanoporous crystalline materials, Magnetic materials |
183. Are LLMs Ready for Real-World Materials Discovery?, Preprint (September 25, 2024)
| Category | Items |
|---|---|
| Datasets | MaScQA, Battery Device QA, MatSciNLP, OpticalTable / OpticalTable-SQA, SustainableConcrete, Cambridge Structural Database (CSD), Publisher machine-readable MatSci corpus (Springer/Elsevier APIs), S2ORC (Semantic Scholar Open Research Corpus), RedPajama (open dataset) |
| Models | GPT, BERT, Transformer, Variational Autoencoder, Diffusion Model, Graph Neural Network, Message Passing Neural Network |
| Tasks | Question Answering, Named Entity Recognition, Information Retrieval, Text Generation, Structured Prediction, Sequence Labeling, Feature Extraction, Image Classification, Clustering |
| Learning Methods | Pre-training, Fine-Tuning, In-Context Learning, Transfer Learning, Few-Shot Learning, Weakly Supervised Learning |
| Performance Highlights | accuracy_overall: 62%, numerical_questions_accuracy: 39%, relative_to_top_humans: ≈50% of top-performing humans, code_generation_accuracy: 71%, table_property_extraction_recall: ≈55% |
| Application Domains | Materials Science (core), Crystallography / inorganic materials design, Chemistry (adjacent domain; synthesis/retrosynthesis), Energy storage (batteries, electrolytes), Cement / concrete (sustainable concrete design), Glass science and optical materials, Robotics / autonomous laboratories (experimental execution), Healthcare / biomedical materials (medical implants), Manufacturing / materials processing |
182. Electronic descriptors for dislocation deformation behavior and intrinsic ductility in bcc high-entropy alloys, Science Advances (September 20, 2024)
| Category | Items |
|---|---|
| Datasets | DFT dislocation core and vibrational dataset for selected bcc HEAs, DFT bulk DOS descriptor dataset (cubic 54-atom cells) across >50 bcc complex concentrated alloys, Surface and unstable stacking fault (USF) energy dataset, Experimental compressive fracture strain dataset (literature compilation) |
| Models | None |
| Tasks | Feature Extraction, Ranking, Clustering, Data Generation |
| Learning Methods | None |
| Performance Highlights | None |
| Application Domains | Materials Science, Computational Materials Design, Alloy Discovery / High-Entropy Alloys, Mechanical Behavior / Deformation Mechanisms |
181. Scalable crystal structure relaxation using an iteration-free deep generative model with uncertainty quantification, Nature Communications (September 17, 2024)
| Category | Items |
|---|---|
| Datasets | X-Mn-O dataset, Materials Project (MP) dataset, C2DB (Computational 2D Materials Database), Layered van der Waals (vdW) crystals dataset, MoS2 defect dataset, DeepMind 2.2 million hypothetical crystals (referenced) |
| Models | Graph Neural Network |
| Tasks | Regression, Data Generation |
| Learning Methods | Supervised Learning, Transfer Learning, Pre-training, Fine-Tuning, Ensemble Learning, Gradient Descent |
| Performance Highlights | X-Mn-O_coordinates_MAE_A: 0.116, X-Mn-O_bond_length_MAE_A: 0.136, X-Mn-O_lattice_MAE_A: 0.063, X-Mn-O_cell_volume_MAE_A3: 3.4, X-Mn-O_match_rate_percent: 84.7, MP_coordinates_MAE_A: 0.066, MP_bond_length_MAE_A: 0.094, MP_lattice_MAE_A: 0.041, MP_cell_volume_MAE_A3: 9.6, C2DB_coordinates_MAE_A_DeepRelax: 0.196, C2DB_bond_length_MAE_A_DeepRelax: 0.268, C2DB_lattice_MAE_A_DeepRelax: 0.085, C2DB_cell_volume_MAE_A3_DeepRelax: 60.2, C2DB_coordinates_MAE_A_DeepRelaxT: 0.185, C2DB_bond_length_MAE_A_DeepRelaxT: 0.165, C2DB_lattice_MAE_A_DeepRelaxT: 0.082, C2DB_cell_volume_MAE_A3_DeepRelaxT: 56.7, X-Mn-O_PAINN_coordinates_MAE_A: 0.159, X-Mn-O_PAINN_bond_length_MAE_A: 0.175, X-Mn-O_PAINN_lattice_MAE_A: 0.066, X-Mn-O_PAINN_cell_volume_MAE_A3: 3.8, X-Mn-O_PAINN_match_rate_percent: 81.2, X-Mn-O_EGNN_coordinates_MAE_A: 0.166, X-Mn-O_EGNN_bond_length_MAE_A: 0.189, X-Mn-O_EGNN_lattice_MAE_A: 0.066, X-Mn-O_EGNN_cell_volume_MAE_A3: 4.2, X-Mn-O_EGNN_match_rate_percent: 77.5, X-Mn-O_Cryslator_coordinates_MAE_A: 0.127, X-Mn-O_Cryslator_cell_volume_MAE_A3: 6.2, X-Mn-O_Cryslator_match_rate_percent: 83.7, Ablation_Dummy_coordinates_MAE_A: 0.314, Ablation_Vanilla_coordinates_MAE_A: 0.155, Ablation_DeepRelax(UCOE)_coordinates_MAE_A: 0.121, Ablation_DeepRelax(BLDU)_coordinates_MAE_A: 0.142, Ablation_DeepRelax_full_coordinates_MAE_A: 0.116, Spearman_correlation_system_uncertainty_vs_error_X-Mn-O: 0.95, Spearman_correlation_system_uncertainty_vs_error_MP: 0.83, Spearman_correlation_system_uncertainty_vs_error_C2DB: 0.88, Energy_MAE_reduction_X-Mn-O_samples_kJ?or_eV?: energy MAE reduced from 32.51 to 5.97 (units: unclear in text but reported as energy MAE), DFT_ionic_steps_reduction_example: Starting DFT from DeepRelax-predicted structures significantly reduces number of ionic steps vs starting from unrelaxed structures (Fig.4f)., Speedup_vs_M3GNet_factor: 100 |
| Application Domains | Materials science (crystal structure relaxation), Computational chemistry / computational materials discovery, 2D materials and defect engineering, High-throughput virtual screening of hypothetical materials |
180. De novo design of high-affinity protein binders with AlphaProteo, Preprint (September 12, 2024)
| Category | Items |
|---|---|
| Datasets | Protein Data Bank (PDB) — structural examples used for training and benchmarking, Distillation set of AlphaFold predictions, Cao et al. de novo binder dataset (retrospective benchmark), Published RFdiffusion designs (downloaded set), This paper: experimental screening dataset (yeast surface display + follow-up expression/KD), Experimental neutralization assay data (SARS-CoV-2) |
| Models | Diffusion Model, Graph Neural Network, Convolutional Neural Network, Transformer |
| Tasks | Synthetic Data Generation, Binary Classification, Regression, Ranking, Clustering |
| Learning Methods | Generative Learning, Pre-training, Knowledge Distillation, Zero-Shot Learning, Supervised Learning, Representation Learning |
| Performance Highlights | experimental_success_rate_BHRF1_percent: 88, experimental_success_rate_SC2RBD_percent: 12, experimental_success_rate_IL-7RA_percent: 25, experimental_success_rate_PD-L1_percent: 15, experimental_success_rate_TrkA_percent: 9, experimental_success_rate_IL-17A_percent: 14, experimental_success_rate_VEGF-A_percent: 33, experimental_success_rate_TNFα_percent: 0, best_per_target_KD_BHRF1_nM: 8.5, best_per_target_KD_SC2RBD_nM: 26, best_per_target_KD_IL-7RA_nM: 0.082, best_per_target_KD_PD-L1_nM: 0.18, best_per_target_KD_TrkA_nM: 0.96, best_per_target_KD_IL-17A_nM: 8.4, best_per_target_KD_VEGF-A_nM: 0.48, overall_best_KD_nM: 0.082, fold_improvement_vs_best_previous_unoptimized_range: 3x to 300x (per abstract/main results), notable_effect: improved filter increased experimental success on SC2RBD and PD-L1 (see Section S2 and Table S2), Example_SC2RBD_success_with_improved_filters_percent (AlphaProteo v1 improved filters): 29, Example_PD-L1_success_with_v2_percent (AlphaProteo v2 on PD-L1): 26.5, AF3-optimized_filter_thresholds: min pae interaction <1.5; ptm binder >0.8; rmsd <2.5 (derived in Section S2.2), retrospective_enrichment: AF3-based filters enriched for experimental success more strongly than AF2-based filters on Cao et al. dataset (Section S2.2, Figure S1C), role: ProteinMPNN used to redesign sequences with low sampling temperature (0.0001) during AF2-based benchmark reproduction, contextual_performance: used as part of AF2 benchmark reproduction pipeline; not reported as a standalone performance metric in paper |
| Application Domains | Protein design / structural biology, Therapeutics (binder design for therapeutic targets such as PD-L1, TrkA, IL-7RA, VEGF-A, IL-17A, TNFα), Virology (SARS-CoV-2 neutralization), Biotechnology / research reagents (ready-to-use binders for imaging, signaling modulation, etc.), Cryo-EM and X-ray structural validation workflows |
179. ChemOS 2.0: An orchestration architecture for chemical self-driving laboratories, Matter (September 04, 2024)
| Category | Items |
|---|---|
| Datasets | DFT calculations (ChemOS 2.0 DFT database uploaded to ioChem-BD), Experimental database (internal ChemOS 2.0 experimental RDBMS), Enumerated BSBCz derivative search space (products of double Suzuki-Miyaura coupling with 38 commercially available dihalides), Frozen ChemOS 2.0 code/version and experimental data (GitHub repository / Zenodo snapshot) |
| Models | Gaussian Process |
| Tasks | Optimization, Experimental Design, Regression |
| Learning Methods | Active Learning |
| Performance Highlights | None |
| Application Domains | Materials discovery, Chemistry (synthetic organic chemistry), Optoelectronic materials / organic laser molecules, Automated experimentation / self-driving laboratories, Computational chemistry (DFT simulations) |
178. Closed-loop transfer enables artificial intelligence to yield chemical knowledge, Nature (September 2024)
| Category | Items |
|---|---|
| Datasets | 2,200-molecule design space (donor–bridge–acceptor combinatorial space), BO-synthesized experimental rounds (Phase I): 30 molecules, Full experimental photostability dataset (CLT campaign), Predicted photostabilities across 2,200 molecules (DFT+RDKit featurizations) |
| Models | Support Vector Machine, Linear Model |
| Tasks | Regression, Optimization, Feature Selection, Feature Extraction, Dimensionality Reduction |
| Learning Methods | Supervised Learning |
| Performance Highlights | LOOV_R2: 0.86, Spearman_R2_on_validation_batches: 0.54, Mann-Whitney_p_value: 0.026, Top7_avg_photostability: 165, Bottom7_avg_photostability: 97, Most_predictive_models_R2_threshold: >0.70, Top5_avg_photostability_improvement: >500%, sampling_fraction: <1.5% of 2,200 space |
| Application Domains | molecular photostability / photodegradation for light-harvesting small molecules, organic electronics (organic photovoltaics, organic light-emitting diodes), dyed polymers and photo-active coatings, solar fuels and photosynthetic system analogues, organic laser emitters (mentioned as further application), stereoselective aluminium complexes for ring-opening polymerization (mentioned as further application) |
177. AI-driven research in pure mathematics and theoretical physics, Nature Reviews Physics (September 2024)
| Category | Items |
|---|---|
| Datasets | arXiv, viXra, Lean MathLib, MathPile, CICY threefolds / Complete Intersection Calabi–Yau (CICY) datasets, Kreuzer–Skarke Calabi–Yau database, Sliding-window binary sequence dataset (synthetic example in Box 2) |
| Models | Decision Tree, Support Vector Machine, Feedforward Neural Network, Multi-Layer Perceptron, Perceptron, Transformer, Random Forest |
| Tasks | Binary Classification, Classification, Image Classification, Language Modeling, Clustering, Optimization, Sequence-to-Sequence, Language Modeling |
| Learning Methods | Supervised Learning, Unsupervised Learning, Reinforcement Learning, Pre-training, Fine-Tuning |
| Performance Highlights | accuracy_first_sequence: 100%, accuracy_second_sequence: about 80%, accuracy_third_sequence: about 50%, accuracy_second_sequence: about 80%, separation_found: qualitative (separation between simple and non-simple finite groups observed; no numeric metric provided), sequence_task_accuracy_first: 100%, sequence_task_accuracy_second: about 80%, algebraic_variety_task_accuracy: >99.9%, algorithm_discovery: discovered faster matrix multiplication algorithm (reported), later superseded by a human-derived algorithm |
| Application Domains | Pure mathematics, Theoretical physics, Algebraic geometry, Number theory, String theory / string landscape, Representation theory and algebraic structures, Combinatorics and graph theory, Knot theory, Quantum field theory, Theoretical cosmology, Symbolic mathematics / automated theorem proving |
175. Machine learning enables the discovery of 2D Invar and anti-Invar monolayers, Nature Communications (August 14, 2024)
| Category | Items |
|---|---|
| Datasets | C2DB (computational 2D materials database) - selected stable subset, Full 2D materials collections (contextual), Subset for ZA-mode contribution analysis, Training/validation splits used in ML experiments |
| Models | Random Forest, Support Vector Machine |
| Tasks | Classification, Regression, Feature Selection, Feature Extraction |
| Learning Methods | Supervised Learning, Ensemble Learning, Bagging, Active Learning |
| Performance Highlights | accuracy: 100% (all training data perfectly classified), alpha500K_regression_RMSE_train: 1.67 × 10^-6 K^-1, alpha500K_regression_RMSE_test: 1.35 × 10^-6 K^-1, alpha500K_regression_R2_train: 0.91, alpha500K_regression_R2_test: 0.93, D_prediction_RMSE: 1.34 eV |
| Application Domains | Materials science, 2D materials, Thermal expansion / thermal management, Nanotechnology / nanoelectronics, Computational materials discovery / high-throughput materials screening |
174. Accurate prediction of protein function using statistics-informed graph networks, Nature Communications (August 04, 2024)
| Category | Items |
|---|---|
| Datasets | PDB-derived GO benchmark (41,896 protein chains), PDB-derived EC benchmark (20,215 protein chains), CAFA3 dataset, Independent hold-out set (RCSB PDB post-2022), UniClust30 multiple sequence alignments (MSAs), UniProt / UniRef50 (ESM-1b pretraining corpora), BioLiP (semi-manually curated ligand-protein interactions), PhiGnet application output on UniProt |
| Models | Graph Convolutional Network, Transformer, Convolutional Neural Network, Multi-Layer Perceptron, Triplet Network, Graph Neural Network |
| Tasks | Multi-label Classification, Multi-class Classification, Sequence Labeling, Representation Learning, Feature Extraction |
| Learning Methods | Supervised Learning, Pre-training, Transfer Learning, Fine-Tuning, Mini-Batch Learning, Representation Learning, Contrastive Learning, End-to-End Learning |
| Performance Highlights | AUPR_overall: 0.70, F_max_overall: 0.80, AUPR_CC: 0.64, F_max_CC: 0.82, AUPR_BP: 0.65, F_max_BP: 0.75, AUPR_MF: 0.80, F_max_MF: 0.81, MCC_average: 0.76, AUPR: 0.89, F_max: 0.88, robust_F_max_at_30%_seqid: 0.61, robust_F_max_at_40%_seqid: 0.72, residue_level_accuracy_on_9_proteins: >=75% (average), qualitative_examples: near-perfect predictions for cPLA2α, Ribokinase, αLA, TmpK, and Ecl18kI, EC_F_max: 0.37, EC_AUPR: 0.21, EC_F_max: 0.69, EC_AUPR: 0.70, EC_F_max: 0.76, EC_AUPR: 0.70, CAFA3_F_max_BP: 0.458, CAFA3_F_max_CC: 0.493, CAFA3_F_max_MF: 0.470, CAFA3_AUPR_BP: 0.378, CAFA3_AUPR_CC: 0.361, CAFA3_AUPR_MF: 0.323 |
| Application Domains | Protein function prediction / bioinformatics, Structural biology (residue-level functional site identification), Genomics / proteomics (large-scale UniProt annotation), Drug discovery and biomedical research (interpretation of functional residues and disease variants), Evolutionary biology (leveraging evolutionary couplings and residue communities) |
173. Accelerated discovery of perovskite solid solutions through automated materials synthesis and characterization, Nature Communications (August 02, 2024)
| Category | Items |
|---|---|
| Datasets | Experimental dataset from ICSD (perovskites and non-perovskites), Candidate pool of disordered compositions, Materials Project ABO3 subset (stable/metastable perovskites), Experimental data and code repository |
| Models | Gradient Boosting Tree, Variational Autoencoder |
| Tasks | Binary Classification, Ranking, Recommendation, Feature Extraction, Representation Learning |
| Learning Methods | Supervised Learning, Unsupervised Learning, Representation Learning, Feature Learning |
| Performance Highlights | accuracy: 94% |
| Application Domains | Materials science (perovskite oxide discovery), Automated materials synthesis (self-driving laboratory / robotic synthesis), High-frequency dielectric characterization (microwave dielectric materials), Wireless communications (tunable devices, antennas) and biosensors (application areas motivating materials discovery) |
172. The power and pitfalls of AlphaFold2 for structure prediction beyond rigid globular proteins, Nature Chemical Biology (August 2024)
| Category | Items |
|---|---|
| Datasets | AlphaFold Protein Structure Database, Protein Data Bank (PDB), Human proteome models (AF2 coverage), McDonald et al. peptide benchmark (588 peptides), Yin et al. heterodimer benchmark (152 heterodimeric complexes), Bryant et al. heterodimer benchmark (dataset used in their study), Terwilliger et al. molecular-replacement benchmark (215 structures), Membrane protein benchmarks (various sets), NMR ensemble datasets (general), SAXS / SANS datasets and Small-Angle Scattering Biological Data Bank derived datasets |
| Models | Transformer, Attention Mechanism |
| Tasks | Structured Prediction, Sequence-to-Sequence, Clustering |
| Learning Methods | Pre-training, Fine-Tuning, Ensemble Learning, Representation Learning, Transfer Learning, Stochastic Learning |
| Performance Highlights | human_proteome_coverage: 98.5% modeled, high_confidence_residue_fraction: ~50% of residues across all proteins predicted with high confidence (cited average), pLDDT_thresholds: pLDDT > 70 interpreted as higher confidence; pLDDT > 90 as very high, peptide_benchmark_size: 588 peptides (McDonald et al.), peptide_prediction_note: AF2 predicts many α-helical and β-hairpin peptide structures with surprising accuracy (no single numeric accuracy given in paper excerpt), heterodimer_success_Yin: 51% success rate (AF2 and AlphaFold2-Multimer on 152 heterodimeric complexes), heterodimer_success_Bryant: 63% success rate (Bryant et al. study), molecular_replacement_success: 187 of 215 structures solved using AlphaFold-guided molecular replacement (Terwilliger et al.), alternative_conformation_sampling_note: modifications (reduced recycles, shallow MSAs, MSA clustering, enabling dropout) allow sampling of alternative conformations (no single numeric accuracy provided), AlphaMissense_note: AlphaMissense provides probability of missense variant pathogenicity; AF2 itself ‘has not been trained or validated for predicting the effect of mutations’ (authors’ caution) |
| Application Domains | Structural biology (protein 3D structure prediction and validation), Proteomics (proteome-scale modeling; human proteome coverage), Integrative structural methods (integration with SAXS, NMR, cryo-EM, X-ray diffraction), Drug discovery / therapeutics (identifying therapeutic candidates, ligand/cofactor modeling), Membrane protein biology (transmembrane protein modeling), Intrinsically disordered proteins (IDPs/IDRs) and conformational ensembles, Peptide biology and peptide–protein interactions, De novo protein design |
171. OpenFold: retraining AlphaFold2 yields new insights into its learning mechanisms and capacity for generalization, Nature Methods (August 2024)
| Category | Items |
|---|---|
| Datasets | OpenProteinSet (replication of AlphaFold2 training set), Protein Data Bank (PDB), Uniclust MSAs, CAMEO validation set, CASP15 domains, CATH-derived domain splits (topologies/architectures/classes), Subsampled training sets (ablation experiments), Rosetta decoy ranking dataset (subset) |
| Models | Transformer, Attention Mechanism, Self-Attention Network, Multi-Head Attention |
| Tasks | Regression, Sequence Labeling, Binary Classification, Ranking |
| Learning Methods | Supervised Learning, Knowledge Distillation, Fine-Tuning, Pre-training, Self-Supervised Learning, Distributed Learning, Ensemble Learning |
| Performance Highlights | OpenFold (mean lDDT-Cα on CAMEO, main comparison): 0.911, AlphaFold2 (mean lDDT-Cα on CAMEO, main comparison): 0.913, OpenFold final replication (after clamping change): 0.902 lDDT-Cα (on CAMEO validation set), Full data model peak (early reported value): 0.83 lDDT-Cα (after 20,000 steps), 10,000-sample subsample after 7,000 steps: exceeded 0.81 lDDT-Cα, 1,000-chain ablation (short run, ~7,000 steps): 0.64 lDDT-Cα, Inference speedup (overall OpenFold vs AlphaFold2): up to 3-4x faster (single A100 GPU), FlashAttention effect on short sequences (<1000 residues): up to 15% additional speedup in OpenFold when applicable, Sequence length robustness: OpenFold runs successfully on sequences and complexes exceeding 4,000 residues; AlphaFold2 crashes beyond ~2,500 residues on single GPU, Secondary structure learning order (qualitative): α-helices learned first, then β-sheets, then less common SSEs (measured by F1 over DSSP categories); final high F1 scores for SSEs, Contact F1 (for fragments / SSEs): improves earlier for shorter helices and narrower sheets; specific numbers are plotted in Fig. 5b and Extended Data Fig. 8 (no single consolidated numeric value in text), Number of models in final ensemble: 10 distinct models (seven snapshots from main run + additional models from branch), Effect: Ensemble used at prediction time to generate alternate structural hypotheses; explicit ensemble metric improvement not numerically summarized in a single value in main text |
| Application Domains | Protein structural biology, Biomolecular modeling (protein complexes, peptide–protein interactions), Evolutionary sequence analysis / MSA-based modeling, RNA structure prediction (discussed as potential application), Spatial reasoning over polymers and arbitrary molecules (structure module / invariant point attention) |
170. Large-scale foundation model on single-cell transcriptomics, Nature Methods (August 2024)
| Category | Items |
|---|---|
| Datasets | Pretraining corpus of human single-cell RNA-seq (scRNA-seq) profiles, Validation split of pretraining data, Read-depth enhancement test set (unseen cells), Zheng68K, Segerstolpe dataset, Baron (pancreatic islet) dataset processed by SAVER, Cancer drug response (CDR) dataset (DeepCDR preprocessed), Single-cell drug response classification dataset (SCAD), Perturbation datasets (Perturb-seq resources), Simulated reference and query dataset; organoid and in vivo data |
| Models | Transformer, Multi-Layer Perceptron, Graph Neural Network, Variational Autoencoder, BERT, GPT, Attention Mechanism |
| Tasks | Regression, Clustering, Multi-class Classification, Binary Classification, Graph Generation, Link Prediction, Representation Learning |
| Learning Methods | Self-Supervised Learning, Pre-training, Transfer Learning, Fine-Tuning, Domain Adaptation, Supervised Learning, Representation Learning, Pre-training |
| Performance Highlights | MAE: about 50% reduction (text: ‘notable reduction of half the MAE’), MRE: about 50% reduction (text: ‘notable reduction of half the … MRE’), PCC: substantial increase (visual reported; specific case examples given in Fig. 2b), PCC_best_case: 0.93 (overall example: Pearson: 0.93, Spearman: 0.87, N = 73), PCC_WZ-1-84: Pearson: 0.94, Spearman: 0.95, N = 8, Leave-one-drug-out_improvement: Top drug PHA-793887: PCC improved from 0.07 to 0.73; example drug zibotentan improved from 0.49 to 0.64, AUCs_scFoundation: 0.84, 0.84, 0.66, 0.68 (four drugs reported in figure), AUCs_baseline: 0.62, 0.56, 0.38, 0.66 (baseline SCAD with raw gene expression), Spearman_correlations: For NVP-TAE684: scFoundation 0.56 vs baseline 0.24; for sorafenib: scFoundation -0.55 vs baseline -0.06, MSE: scFoundation-based GEARS achieved lower averaged MSE compared with baseline GEARS and CPA across tested perturbation datasets (numerical MSEs reported in figures / Supplementary), Averaged_MSE_two-gene_0/2_unseen: lowest averaged MSE for scFoundation-based model (text statement), PCC_magnitude_score: scFoundation Pearson: 0.18; baseline Pearson: 0.01 (Fig. 5e), macro_F1: scFoundation achieved the highest macro F1 score on both Zheng68K and Segerstolpe datasets (exact numeric values provided in Supplementary Table 4), improvement_on_rare_types: improved performance on rare cell types such as CD4+ T helper 2 and CD34+, NMI/ARI/SIL: scFoundation outperformed baseline downsampled data and other imputation methods (scImpute, scVI, SAVER, MAGIC) across these clustering metrics (see Fig. 2c and 2f), SIL_on_Zheng68K: scFoundation had a higher silhouette coefficient compared to scVI and raw data (text: ‘scFoundation had a higher SIL score’) |
| Application Domains | single-cell transcriptomics / computational biology, cancer pharmacogenomics (drug response prediction), single-cell perturbation analysis (Perturb-seq), cell type annotation and atlas construction, gene module discovery and gene regulatory network inference, bioinformatics data integration and batch correction |
169. Sequential closed-loop Bayesian optimization as a guide for organic molecular metallophotocatalyst formulation discovery, Nature Chemistry (August 2024)
| Category | Items |
|---|---|
| Datasets | Virtual library of 560 CNP molecules, First experimental CNP dataset (BO-synthesized), Expanded experimental CNP dataset (BO + diversity baseline), Predicted set of 100 new CNPs, Reaction-condition space (encoded), Experimental reaction-condition dataset (BO + initial) |
| Models | Gaussian Process, Radial Basis Function Network |
| Tasks | Optimization, Regression, Experimental Design, Dimensionality Reduction, Feature Extraction |
| Learning Methods | Supervised Learning, Active Learning, Batch Learning |
| Performance Highlights | max_yield_first_bo_campaign: 67%, samples_evaluated_first_campaign: 55 of 560 CNPs, max_yield_reaction_condition_optimization: 88%, samples_evaluated_reaction_conditions: 107 of 4,500 possible conditions, BO_vs_random_max_yield: 88% (BO) vs 75% (random baseline), high_activity_fraction_BO: 39 of 88 (44%) samples had yield >67%, high_activity_fraction_random: 2 of 44 (4.5%) samples had yield >=67%, prediction_accuracy_examples: Predicted yields for new CNPs matched experimental yields for three synthesized candidates (CNP-561, CNP-565, CNP-577) ‘close to their predicted values’ (no numeric error given in text) |
| Application Domains | organic chemistry, photoredox catalysis, metallaphotocatalysis (photoredox + nickel dual catalysis), catalyst discovery, reaction-condition optimization, closed-loop experiment design / autonomous/algorithm-guided experimentation |
168. Compositional design of multicomponent alloys using reinforcement learning, Acta Materialia (August 01, 2024)
| Category | Items |
|---|---|
| Datasets | 112 composition-enthalpy dataset (initial training data), Candidate search space of multicomponent alloys, Experimental dataset synthesized in this work, Nine large-enthalpy samples (thermal diffusivity measurements), Synthetic benchmark functions (Ackley, Rastrigin, Levy) |
| Models | Gaussian Process, Gradient Boosting Tree, Multi-Layer Perceptron |
| Tasks | Regression, Decision Making, Optimization, Experimental Design, Feature Selection / Feature Extraction |
| Learning Methods | Reinforcement Learning, Supervised Learning, Active Learning, Pre-training, Mini-Batch Learning, Evolutionary Learning |
| Performance Highlights | training_size: 112 samples, example_prediction_vs_experiment_error_J_per_g: -31.94 (predicted) vs -31.1 (experimental) for Ti53Ni47 (error ~0.84 J/g), diagnostics_reported: R2 increased and RMSE decreased as iterations proceed (exact numerical R2/RMSE values shown in Fig. 2b in paper), training_episodes_to_converge: ≈3,000 (converges in less than 4,000 episodes across seeds), surrogate_calls: agent needs less than 4,000 ×5 surrogate model interactions to propose compositions with maximum cumulative rewards (calls per episode: 5), final_experimental_result: Ti27.2Ni47Hf13.8Zr12 with transformation enthalpy ΔH = -37.1 J/g (corrected to -39.0 J/g after calibration), success_rate_experiment: 33 out of 40 synthesized compositions exhibited martensitic transformation, selected_features: Pettifer chemical scale, valence electron numbers of average atomic number, difference of atomic radii, configurational entropy, contextual_note: GB used to rank feature importance prior to GP training, pretraining_effect: Pretrained agent accesses regions of higher reward values more frequently and learns high-rewarding states with greater probability than agent without pretraining (t-SNE, entropy reduction reported in supplemental figures), scaling_reduction: Interactions with surrogate reduced from O(N) in BGO to ~ O(5255 × N^0.06) for ordered-substitution case (and ~ O(715 × N^0.176) if order is random) estimated by fitting required interactions vs N, black-box_benchmarks: RL outperforms BGO and GA for Ackley, Rastrigin, Levy functions across dimensionalities up to 10 when function call budget is large (3,000); with small budgets (100) RL underperforms, relative_performance_with_budget_3000: GA and BGO are outperformed by RL on most tested functions/dimensionalities up to 10, relative_performance_with_budget_100: With fewer experimental iterations (100), BGO and GA outperform RL for certain problems (e.g., Rastrigin and higher-dimensional Ackley) |
| Application Domains | Materials science (alloy design), Phase change materials (PCMs), Shape memory alloys (SMAs), Thermal energy storage / thermal management, Autonomous experimentation / closed-loop experimental design, Black-box optimization and algorithm benchmarking |
167. Large Language Models for Inorganic Synthesis Predictions, Journal of the American Chemical Society (July 24, 2024)
| Category | Items | |||
|---|---|---|---|---|
| Datasets | Materials Project + Open Quantum Materials Database (unique inorganic compositions), Subset used for LLM fine-tuning (synthesizability), Text-mined inorganic materials synthesis recipes (Kononova et al. dataset, curated) |
|||
| Models | GPT, Graph Neural Network |
|||
| Tasks | Binary Classification, Multi-label Classification |
|||
| Learning Methods | Fine-Tuning, Pre-training, Positive-Unlabeled Learning, Semi-Supervised Learning |
|||
| Performance Highlights | alpha_estimate: 0.088, probability_threshold_for_recalibration_GPT-3.5(FT): 0.761, note_p(P |
U)_at_0.5_threshold_for_stoi-CGNF_and_GPT-3.5(FT): 15% of unlabeled data predicted as positive (p(P | U)) at 0.5 threshold, behavior: very high recall but very low precision when using 0.5 threshold; predicted positive class for many inputs, probability_threshold_for_recalibration_GPT-3.5(pretrained): 0.977, probability_threshold_for_recalibration_GPT-4(pretrained): 0.963, probability_threshold_for_recalibration_stoi-CGNF: 0.723, note: At 0.5-threshold, stoi-CGNF and GPT-3.5(FT) predicted 15% of unlabeled as positive (p(P |
U)) which is inconsistent with α=0.088, Top-5_accuracy_GPT-3.5(FT): 86.0%, Top-1_accuracy_GPT-3.5(FT): comparable to Elemwise (exact Top-1 numeric in Fig.2/Table Sx in SI), fine-tune_time_cost: <90 min and <11 USD per GPT-3.5 model (as of 04/2024), ensemble_Top-5_accuracy_after_GPT-4_feasibility_filter: 87.6%, Top-5_accuracy_combination_model_retaining_first_5_unique_allowed_reactions: 90.9%, combined_Top-5_union_correct_rate: 93% (Combining Top-5 predictions of both models would predict the correct precursors for 93% of target compounds) |
| Application Domains | inorganic chemistry, solid-state chemistry, materials science, synthesis planning / experimental synthesis, computational materials discovery |
166. Autonomous chemistry: Navigating self-driving labs in chemical and material sciences, Matter (July 03, 2024)
| Category | Items |
|---|---|
| Datasets | literature reaction / chemistry datasets (general), Adam platform measurements (bioinformatics / enzyme search), A-lab generated inorganic compounds dataset, High-throughput experimental datasets (HPLC, NMR, IR, mass spec outputs) |
| Models | Gaussian Process, Decision Tree, Random Forest, Multi-Layer Perceptron, Transformer |
| Tasks | Optimization, Optimization, Optimization, Data Generation, Optimization |
| Learning Methods | Gradient Descent, Reinforcement Learning, Evolutionary Learning, Multi-Task Learning, Active Learning, Supervised Learning |
| Performance Highlights | None |
| Application Domains | synthetic chemistry / reaction optimization, materials science / material property optimization, analytical chemistry (e.g., chromatography method optimization), drug discovery / compound discovery, biological discovery (gene/enzyme identification), automation/robotics integration and lab orchestration |
165. Has generative artificial intelligence solved inverse materials design?, Matter (July 03, 2024)
| Category | Items |
|---|---|
| Datasets | MatterGen pretraining dataset (~1 million unique bulk crystal structures), public materials datasets (general), stable materials subset (used by CDVAE) |
| Models | Variational Autoencoder, Generative Adversarial Network, Denoising Diffusion Probabilistic Model, Transformer, Graph Neural Network, Graph Convolutional Network, Convolutional Neural Network, Conditional GAN, GPT, DALL-E |
| Tasks | Synthetic Data Generation, Graph Generation, Text Generation, Language Modeling, Regression, Optimization, Distribution Estimation, Synthetic Data Generation |
| Learning Methods | Unsupervised Learning, Self-Supervised Learning, Adversarial Training, Pre-training, Fine-Tuning, Representation Learning, Adversarial Training |
| Performance Highlights | reconstruction_loss: not numerically specified (qualitative: FTCP suffers heavy reconstruction losses; WYCRYST demonstrates lower reconstruction losses than FTCP), validity: WYCRYST demonstrates higher structure validity than FTCP (qualitative), precision: VAE showed slightly higher precision and was easier to train than a Wasserstein GAN in one elpasolite composition study (qualitative, no numeric value), structure_validity: diffusion models outperform GANs on structure validity (qualitative), distribution_quality: learned distribution of crystal structure parameters better for diffusion (C RYSTENS) than GANs (qualitative), low_energy_targeting: CRYSTAL-LLM (fine-tuned LLAMA-2) outperforms early diffusion-based models (e.g., CDVAE) in targeting low-energy configurations (qualitative) |
| Application Domains | Inverse materials design for inorganic crystalline materials, Crystal structure generation and prediction, Property-targeted materials design (e.g., band gap, bulk modulus), Porous materials / zeolite design, Surface structure discovery (oxide formation on surfaces), Multi-component alloys and composition design, Bulk metallic glasses design, Thin-film synthesis / processing parameter design, General materials discovery workflows (screening + generative proposals) |
164. Promising directions of machine learning for partial differential equations, Nature Computational Science (July 2024)
| Category | Items |
|---|---|
| Datasets | 2D simulations of fluid flow past a circular cylinder, Large-scale mesoscale ocean model outputs (LES closure training data), High-fidelity particle-in-cell simulations, Kolmogorov 2D flow dataset (Kochkov et al.) / turbulent flow fields, Forced turbulence training dataset (illustrated in Fig. 5), PDEBench (benchmark problems) |
| Models | Autoencoder, Convolutional Neural Network, Long Short-Term Memory, Transformer, Graph Neural Network, Gaussian Process, Support Vector Machine, ResNet, Feedforward Neural Network, Multi-Layer Perceptron |
| Tasks | Regression, Dimensionality Reduction, Time Series Forecasting, Image Super-Resolution, Feature Extraction, Clustering, Optimization |
| Learning Methods | Supervised Learning, Unsupervised Learning, Ensemble Learning, Representation Learning, Transfer Learning, Bayesian / Probabilistic Methods (mapped to available list via Gaussian Process), End-to-End Learning |
| Performance Highlights | Reynolds_number_identification_error: within 1% of true value, speedup: ≈86× (figure text) ; described as ‘two orders of magnitude acceleration’ in accuracy vs computational cost comparison |
| Application Domains | Fluid dynamics / turbulence modeling, Climate science / oceanography, Plasma physics, Weather forecasting, Neuroscience (spatiotemporal brain data), Epidemiology, Materials science, Biology (collective dynamics, bacterial colonies, organized biological matter) |
163. Prediction of DNA origami shape using graph neural network, Nature Materials (July 2024)
| Category | Items |
|---|---|
| Datasets | Graph dataset of DNA origami designs (labelled), Augmented unlabelled designs (automatically generated variants), Collected designs (Methods section), Supramolecular assemblies (hierarchical graphs) |
| Models | Graph Neural Network, Gated Recurrent Unit, Transformer, Multi-Head Attention, Ensemble Learning, Residual Network (as residual blocks) |
| Tasks | Regression, Structured Prediction, Graph Generation, Optimization, Data Augmentation |
| Learning Methods | Supervised Learning, Unsupervised Learning, Pre-training, Fine-Tuning, Ensemble Learning, Data Augmentation, Gradient Descent, Backpropagation, Evolutionary Learning, Meta-Learning |
| Performance Highlights | RMSD_mean: 5.56 nm, Orientation_Score_mean: 0.92, RMSD_mean: 6.11 nm, Orientation_Score_mean: 0.92, RMSD_mean: 7.50 nm, Orientation_Score_mean: 0.88, RMSD_mean: 10.10 nm, Orientation_Score_mean: 0.73, RMSD_mean: 3.31 nm, Orientation_Score_mean: 0.95, Inference_time_mean: 0.98 s, Total_energy_ratio_initial_to_GT: 182.3 -> predicted 4.1, Computed_radii_vs_experiment_tetrahedron: 131 nm (pred) vs 125 nm (exp), Computed_radii_vs_experiment_hexahedron: 151 nm (pred) vs 150 nm (exp), Computed_radii_vs_experiment_dodecahedron: 217 nm (pred) vs 210 nm (exp), Predicted_twist_angle: 3.5° (pred) vs ~4.0° (exp), Computation_time_max: up to 66 h (CPU), Number_of_predictions_for_optimization: 5,000 to 100,000 predictions per optimization, Optimization_time_range: 7.8 min to 10.6 h (using pretrained DGNN), SNUPI_alternative_time_estimate: would take hundreds to tens of thousands of hours for same optimization |
| Application Domains | DNA origami / structural DNA nanotechnology, Molecular architecture design, Supramolecular assembly analysis, Inverse design and automated design optimization of nanoscale DNA structures, Rapid virtual prototyping of DNA nanostructures |
162. From bulk effective mass to 2D carrier mobility accurate prediction via adversarial transfer learning, Nature Communications (June 25, 2024)
| Category | Items |
|---|---|
| Datasets | Collected 2D carrier mobility dataset (training/testing), Bulk effective mass data (source-domain properties), C2DB + 2DMatPedia (prediction / screening set), DFT validation data (effective-mass approximation and deformation potential approximation) |
| Models | Multi-Layer Perceptron, XGBoost |
| Tasks | Regression, Regression, Dimensionality Reduction, Feature Extraction, Ranking, Feature Selection |
| Learning Methods | Transfer Learning, Adversarial Training, Domain Adaptation, Backpropagation, Supervised Learning, Ensemble Learning, Boosting, Representation Learning, Pre-training |
| Performance Highlights | R2_electron: 0.88, R2_hole: 0.90, MAE_electron: 0.19, MAE_hole: 0.19, R2_electron: 0.89, MAE_electron: 0.11, R2_hole: 0.89, MAE_hole: 0.13, R2_electron: 0.95, MAE_electron: 0.12, R2_hole: 0.89, MAE_hole: 0.28, R2_overall: >0.82, MAE_overall: <0.22 |
| Application Domains | Materials discovery, 2D materials (semiconductor) design, Electronic materials and device engineering (carrier mobility prediction for semiconductors / transistor scaling), High-throughput screening and materials screening, Catalysis and photovoltaic materials (potential applications of screened materials) |
161. LLMatDesign: Autonomous Materials Discovery with Large Language Models, Preprint (June 19, 2024)
| Category | Items |
|---|---|
| Datasets | Materials Project (structures used to curate datasets and starting materials), mpgap (MatBench), mpform (MatBench), MLFF training dataset (curated), Experimental runs: 10 starting materials (random selection) |
| Models | GPT, Transformer, Attention Mechanism |
| Tasks | Regression, Optimization, Language Modeling, Text Generation, Decision Making, Data Generation |
| Learning Methods | Zero-Shot Learning, Pre-training, Supervised Learning, Prompt Learning |
| Performance Highlights | average_number_of_modifications_to_reach_band_gap_target: 10.8, average_final_band_gap_eV: 1.39, average_formation_energy_eV_per_atom (ML surrogate runs): -1.97, minimum_formation_energy_eV_per_atom (ML surrogate runs): -2.72, average_number_of_modifications_with_history: 13.7, average_number_of_modifications_historyless: 14.8, MLFF_training_dataset_size: 187687, MLFF_training_epochs: 400, MLPP_band_gap_dataset_size: 106113, MLPP_formation_energy_dataset_size: 132752, MLPP_training_epochs_each: 200, average_number_of_modifications_GPT-4o_Refined: 8.69, average_number_of_modifications_Persona: 9.11, average_number_of_modifications_history_baseline: 10.8, average_number_of_modifications_with_history_without_self-reflection: 23.4, average_number_of_modifications_with_history_with_self-reflection: 10.8, average_number_of_modifications_historyless: 26.6, random_baseline_average_number_of_modifications: 27.4, random_baseline_average_final_band_gap_eV: 1.06, DFT_validation_average_formation_energy_per_atom_GPT-4o_with_history: -2.31, DFT_validation_average_formation_energy_per_atom_random: -1.51, DFT_job_success_rate_GPT-4o_with_history_percent: 73.3, DFT_job_success_rate_random_percent: 40.0, constraint_do_not_use_Ba_or_Ca_percent_compliant: 100, constraint_do_not_modify_Sr_percent_compliant: 100, constraint_do_not_have_more_than_4_distinct_elements_percent_compliant: 99.02 |
| Application Domains | Materials Science, Computational Chemistry, Materials Discovery / Inorganic Crystal Design, Photovoltaics (band gap target selection example), Autonomous Laboratories / Self-driving labs, Scientific Decision Making (LLM-assisted) |
160. Deep learning probability flows and entropy production rates in active matter, Proceedings of the National Academy of Sciences (June 18, 2024)
| Category | Items |
|---|---|
| Datasets | N=2 illustrative system (torus), N=64 swimmers in a harmonic trap, N=4,096 MIPS training dataset (packing fraction φ = 0.5), Transfer / test datasets: N=8,192; N=16,384; N=32,768 (various packing fractions φ=0.01..0.9), Datasets for training/optimization (general) |
| Models | Transformer, Multi-Layer Perceptron, Attention Mechanism, Denoising Diffusion Probabilistic Model |
| Tasks | Distribution Estimation, Feature Extraction, Representation Learning |
| Learning Methods | Unsupervised Learning, Self-Supervised Learning, Transfer Learning, Stochastic Gradient Descent, Mini-Batch Learning, Online Learning, Representation Learning |
| Performance Highlights | generalization_system_size: predictive transfer from N=4,096 training to N up to 32,768 without retraining, packing_fraction_transfer_range: predictions consistent for packing fraction φ from 0.01 to 0.9 (evaluated on N=8,192), qualitative_accuracy_checks: convergence verified via FPE residual and stationarity condition Eq. 13; authors report ‘high accuracy’ though no numeric values provided |
| Application Domains | active matter, statistical mechanics, stochastic thermodynamics, scientific computing / computational physics, many-body nonequilibrium systems |
159. Generative learning facilitated discovery of high-entropy ceramic dielectrics for capacitive energy storage, Nature Communications (June 10, 2024)
| Category | Items |
|---|---|
| Datasets | 77 sets of experimental results (initial dataset), Generated candidate dataset (GM output), Phase-field simulation results (P-E loops and energy density vs Sconfig), Augmented compositional search space (sampling discretization) |
| Models | Encoder-Decoder, Multi-Layer Perceptron, LightGBM, Gaussian Mixture Model, Encoder-Decoder |
| Tasks | Generative Learning, Unsupervised Learning, Classification, Binary Classification, Regression, Data Augmentation, Dimensionality Reduction, Representation Learning, Sampling / Monte Carlo Learning, Optimization, Experimental Design, Feature Extraction |
| Learning Methods | Generative Learning, Unsupervised Learning, Supervised Learning, Monte Carlo Learning, Active Learning, Representation Learning, Dimensionality Reduction |
| Performance Highlights | generated_candidates: 2144, predicted_Ue_threshold: >65 J cm−3, candidates_sampled: 2144 (from latent space region), top_selected_for_experiment: 5 compositions (C-1…C-5), generated_candidates_with_predicted_Ue>65: 2144 (from regression/classifier pipeline), downstream_experimental_success: 5 targeted experiments selected; best experimental Ue = 156 J cm−3 (C-3), use_case: filtering generated compositions to a candidate pool (Ue > 65 J cm−3 predicted), numeric_accuracy: not reported, visualized_components: latent z projected by PCA (Fig. 2b), observations: generated candidates (purple) cluster in latent space; five chosen compositions located in middle dense region |
| Application Domains | Materials Science, Dielectric Materials / Dielectric Capacitors, High-Entropy Ceramics, Capacitive Energy Storage, Thin Film Ceramics |
158. Machine learning-guided realization of full-color high-quantum-yield carbon quantum dots, Nature Communications (June 06, 2024)
| Category | Items |
|---|---|
| Datasets | CQD synthesis dataset (initial + augmented) |
| Models | XGBoost, Gradient Boosting Tree, Decision Tree |
| Tasks | Regression, Optimization, Experimental Design, Hyperparameter Optimization, Feature Selection, Dimensionality Reduction |
| Learning Methods | Supervised Learning, Ensemble Learning, Active Learning |
| Performance Highlights | MSE_PLQY_initial: 0.45, MSE_PLQY_after_4_iterations: ≈0.15, MSE_PLQY_stabilized: ≈0.1, MSE_PL_wavelength: <0.1 (throughout iterations), Objective_utility_improvement: from 39.27% to 75.44 (reported improvement range), PLQY_all_colors: >60% (peak PLQY for all seven colors exceeds 60%), Notable_color_PLQYs: cyan 94% (max), some colors near 100% (cyan, green, orange); several colors approach 70% (purple, blue, red), Iterations_to_meet_goal: 20 iterations (40 guided experiments) to achieve PLQY >50% across all seven colors, Total_experiments_for_final_result: 63 experiments, General_model_note: Used as the algorithmic family via XGBoost; specific performance reported under XGBoost entries |
| Application Domains | Materials Science, Nanochemistry, Luminescent materials / Optoelectronics, LEDs, Bioimaging / Life medicine, Solar cells |
157. Closed-Loop Multi-Objective Optimization for Cu–Sb–S Photo-Electrocatalytic Materials’ Discovery, Advanced Materials (June 04, 2024)
| Category | Items |
|---|---|
| Datasets | Cu–Sb–S HTE experimental dataset (this work), Initial sampling prior (Latin hypercube sampling runs), Materials Project phase diagram (Cu–Sb–S subspaces) |
| Models | Gaussian Process |
| Tasks | Regression, Optimization, Feature Selection, Multi-objective Classification |
| Learning Methods | Active Learning, Batch Learning |
| Performance Highlights | RMSE_Y: 0.05, RMSE_bandgap: 0.19, RMSE_Cu1+/Cu_ratio: 0.17, GPRU_RMSE: high (not numerical; model uncertainty remained high for uniformity), R2_improvements: R2 scores improved over iterations (not all numerical values specified; improvements noted from iterations 2–4 and 6–8), photocurrent_optimum: -186 μA cm^-2 at 0 V vs RHE, photocurrent_baseline: -86 μA cm^-2 (batch 1), relative_improvement: 2.3x |
| Application Domains | photo-electrochemical water splitting (photo-electrocatalysis / photocathode discovery), materials discovery, high-throughput experimentation (automated synthesis and characterization), closed-loop autonomous experimentation |
156. ChatMOF: an artificial intelligence system for predicting and generating metal-organic frameworks using large language models, Nature Communications (June 03, 2024)
| Category | Items |
|---|---|
| Datasets | CoREMOF (Computation-ready experimental MOFs), QMOF, MOFkey, DigiMOF, hMOF / hypothetical MOFs (pre-training corpus), Fine-tuned_MOFTransformer_model (released), Database_for_ChatMOF (hMOF used in genetic algorithm) |
| Models | GPT, Transformer, Generative Adversarial Network, Diffusion Model, Variational Autoencoder |
| Tasks | Information Retrieval, Regression, Graph Generation, Optimization, Ranking, Data Generation |
| Learning Methods | Pre-training, Fine-Tuning, Transfer Learning, Prompt Learning, Few-Shot Learning, Zero-Shot Learning, Supervised Learning, Evolutionary Learning, Reinforcement Learning |
| Performance Highlights | accuracy: 96.9%, token_limit_exceeded_counts: 4/100 (search task), accuracy: 95.7%, token_limit_exceeded_counts: 6/100 (prediction task), accuracy: 87.5%, token_limit_exceeded_counts: 2/10 (generation task), parent-child_overlap_percentage: ~30% overlap (GPT-4 generated children vs parents), example_predicted_accessible_surface_area: 6411.28 m^2/g (predicted), calculated_accessible_surface_area_after_optimization: 7647.62 m^2/g, example_predicted_H2_uptake: 499.998 cm^3/cm^3 (predicted), calculated_H2_uptake_after_optimization: 495.823 cm^3/cm^3, example_predicted_hydrogen_diffusivity: 0.0030176841738998412 cm^2/s (BAZGAM_clean), search_accuracy: 95% (GPT-3.5-turbo, excluding token-limit instances), prediction_accuracy: 91% (GPT-3.5-turbo, excluding token-limit instances), generation_accuracy: 77.8% (GPT-3.5-turbo, excluding token-limit instances), note_on_16k_variant: GPT-3.5-turbo-16k (higher max tokens) did not significantly reduce token-limit errors due to suboptimal code generation, example_generation_convergence: Accessible surface area population average increased from ~3748 m^2/g (initial) to 5554 m^2/g by the 3rd generation, example_target_match: H2 uptake target 500 cm^3/cm^3 achieved: predicted 499.998 cm^3/cm^3, calculated after optimization 495.823 cm^3/cm^3, ranking: Generated rtl+N535+N234 structure’s calculated ASA ranks third-highest in CoREMOF |
| Application Domains | Materials science, Metal–organic frameworks (MOFs) research, Computational materials discovery, Chemistry (porous materials / gas adsorption), Materials informatics / AI for science |
154. Three million images and morphological profiles of cells treated with matched chemical and genetic perturbations, Nature Methods (June 2024)
| Category | Items |
|---|---|
| Datasets | CPJUMP1, Drug Repurposing Hub (subset used to select compounds), RxRx3 (mentioned for comparison), JUMP Cell Painting dataset (larger consortium dataset, referenced) |
| Models | Convolutional Neural Network |
| Tasks | Information Retrieval, Ranking, Anomaly Detection, Clustering, Dimensionality Reduction, Feature Extraction, Image Generation, Style Transfer |
| Learning Methods | Supervised Learning, Representation Learning, Multi-View Learning, Transfer Learning, Domain Adaptation |
| Performance Highlights | training_samples: 7,077 single cells, input_feature_dimension: 3,019 features (reduced non-redundant CellProfiler features), output_image_size: 128 x 128 x 5 channels, evaluation_metrics: Average Precision (AP), mean Average Precision (mAP), fraction retrieved (FR = fraction of perturbations with q < 0.05), statistical_testing: Permutation testing (100,000 shuffles) to assign P values to AP, Benjamini–Hochberg FDR to obtain q-values; similarity metric: cosine similarity (or absolute cosine similarity when both positive and negative correlations considered matches), metric: Average Precision (AP) per perturbation; mean Average Precision (mAP) summarized per task; fraction retrieved (FR = fraction perturbations with q < 0.05), stat_test: Permutation test (100,000 shuffles) to obtain P for each AP, then Benjamini–Hochberg FDR to compute q-values, metric: Cosine similarity distributions and counts beyond percentile thresholds vs null; Fisher’s exact test for directionality, reported_counts: n = 3,728 biologically independent ORF and CRISPR reagents (for ORF vs CRISPR comparison); n = 1,864 independent pairs of compounds and genetic perturbations for compound–genetic comparisons |
| Application Domains | Cell biology (cell morphology profiling), Drug discovery and chemical biology (compound mechanism-of-action identification, virtual screening), Functional genomics (gene function via genetic perturbations), Bioimage analysis / high-content screening, Representation learning and machine learning research (benchmarking new methods on biological imaging data) |
153. Machine intelligence-accelerated discovery of all-natural plastic substitutes, Nature Nanotechnology (June 2024)
| Category | Items |
|---|---|
| Datasets | Initial robot-prepared dataset (boundary definition), Active-learning experimental dataset (stagewise fabricated samples), Combined real + virtual training dataset (after data augmentation), SVM classifier test set, Model expansion experiments (chitosan incorporation), Model-predicted/combinatorial design space outputs, Public data/code repository |
| Models | Support Vector Machine, Multi-Layer Perceptron, Linear Model, Decision Tree, Gradient Boosting Tree, Random Forest |
| Tasks | Multi-class Classification, Regression, Optimization, Clustering, Data Augmentation, Model Interpretation (not a listed ML task; included because performed) |
| Learning Methods | Supervised Learning, Active Learning, Ensemble Learning, Data Augmentation, Cross-Validation (fivefold) |
| Performance Highlights | accuracy: 94.3%, A-grade success_rate: >94%, mean_relative_error_(MRE): ≈17% (champion model after 14 active learning loops), ANN_without_data_augmentation_MRE: >55%, model_expansion_MRE_change: from 107% to 21% after three expansion loops, Not_directly_applicable: committee prediction variance used in acquisition function (A_score = L_hat × sigma_hat), MRE_relative_performance: All had higher MREs than ANN (exact values not reported in main text), context_note: Compared to ANN, other models yielded larger MREs (Fig. 2e summary), predicted_and_experimental_strengths: Model-suggested MMT-rich sample: average σu = 114 ± 18 MPa (n=5); CNF-rich sample: average σu = 98 ± 7 MPa (n=5); after two-step treatments: improved σu to 468.6 ± 52.6 MPa (max 520.7 MPa) and 463.0 ± 35.7 MPa (max 521.0 MPa), context_note: Experimental validation of model-suggested high-strength compositions |
| Application Domains | Materials science / nanocomposite materials, Biodegradable plastics / sustainable materials, Polymer engineering / packaging materials, Structural materials (high-strength bio-based materials), Robotics-assisted experimentation / autonomous materials discovery, Materials informatics / ML-driven materials design |
152. Machine learning-aided generative molecular design, Nature Machine Intelligence (June 2024)
| Category | Items |
|---|---|
| Datasets | ChEMBL, GuacaMol, MOSES, Reaxys (reaction corpus used for SCScore), Ultralarge make-on-demand virtual libraries (examples: 11 billion synthon-based, VirtualFlow 69 billion), Chemistry42 (proprietary platform/data), AlphaFold-predicted protein structures (used as structural input), Benchmarks and oracle collections (sample-efficiency benchmark referenced) |
| Models | Variational Autoencoder, Generative Adversarial Network, Normalizing Flow, Diffusion Model, Recurrent Neural Network, Long Short-Term Memory, Gated Recurrent Unit, Transformer, Graph Neural Network, Seq2Seq, Gradient Boosting Tree, Gaussian Process, Autoencoder, Variational Autoencoder, Denoising Diffusion Probabilistic Model, Genetic Algorithm, Reinforcement Learning, Markov chain Monte Carlo |
| Tasks | Graph Generation, Sequence-to-Sequence, Synthetic Data Generation, Optimization, Regression, Classification, Graph Classification, Data Generation |
| Learning Methods | Reinforcement Learning, Fine-Tuning, Pre-training, Bayesian Optimization, Evolutionary Learning, Monte Carlo Learning, Gradient Descent, Active Learning, Contrastive Learning, Transfer Learning, Few-Shot Learning, Representation Learning |
| Performance Highlights | hit_rate: 4/5 (80%), outcome: nM agonist (RXR) reported, hit_rate: 2/4 (50%), outcome: μM agonist (RXR), hit_rate: 7/7 (100%), outcome: nM inhibitor (JAK1), hit_rate: 2/6 (33%), outcome: nM inhibitor (Nurr1γ), hit_rate: 9/43 (21%), outcome: nM inhibitor (CDK8), hit_rate: 0/1 (0%), outcome: μM inhibitor (Bacteria), hit_rate: 2/2 (100%), outcome: nM inhibitor (DDR1), hit_rate: 1/1 (100%), outcome: nM inhibitor (TBK1), hit_rate: 17/23 (74%), outcome: cnM inhibitor (CDK2) reported, hit_rate: 2/2 (100%), outcome: μM agonist (PPARγ), outcome: design, synthesis and experimental validation of DDR1 inhibitor within 21 days (nanomolar or notable potency reported in ref. 19), hit_rate: 4/6 (67%), outcome: nM inhibitor (DDR1) reported in ref. 108 and Table 4, hit_rate: 7/7 (100%), outcome: nM inhibitor (CDK2) reported in 2024 (ref.173) - lead optimization |
| Application Domains | Drug discovery, Small-molecule design, Structure-based drug design (SBDD), Ligand-based drug design (LBDD), Medicinal chemistry, Pharmacology, Antibiotic discovery, Protein–ligand binding and docking, Automated synthesis / self-driving labs, Materials design (suggested potential extensions: polysaccharides, proteins, nucleic acids, crystals, polymers) |
151. High-Entropy Photothermal Materials, Advanced Materials (June 2024)
| Category | Items |
|---|---|
| Datasets | co-sputtered material libraries (combinatorial material libraries), comprehensive databases of material properties (unnamed) |
| Models | None |
| Tasks | Experimental Design |
| Learning Methods | Supervised Learning, Representation Learning |
| Performance Highlights | None |
| Application Domains | Solar water evaporation, Personal thermal management (warming textiles), Solar thermoelectric generation, Photocatalysis (CO2 reduction, water splitting), Photothermal catalysis (biomass conversion, CO production), Biomedical applications (photothermal therapy, antibacterial/antibiofilm), Broadband solar absorbers, Spectrally selective absorbers |
150. Diffusion-based deep learning method for augmenting ultrastructural imaging and volume electron microscopy, Nature Communications (June 01, 2024)
| Category | Items |
|---|---|
| Datasets | In-house mouse brain cortex dataset (denoising), In-house mouse brain cortex dataset (super-resolution), In-house mouse liver, heart, bone marrow, cultured HeLa cell images, OpenOrganelle mouse liver dataset (jrc_mus-liver), OpenOrganelle mouse kidney dataset (jrc_mus-kidney), OpenOrganelle T-cell dataset (jrc_ctl-id8-2), EPFL mouse brain dataset, MICrONS multi-area dataset, FANC dataset, MANC dataset (downsampled) |
| Models | Denoising Diffusion Probabilistic Model, U-Net |
| Tasks | Image Denoising, Image Super-Resolution, Image-to-Image Translation, Image Generation, Semantic Segmentation |
| Learning Methods | Supervised Learning, Self-Supervised Learning, Transfer Learning, Fine-Tuning, Ensemble Learning, Pre-training |
| Performance Highlights | LPIPS: EMDiffuse-n outperformed baselines (lower LPIPS values across test set; test points n=960), FSIM: EMDiffuse-n outperformed baselines (higher FSIM), Resolution Ratio: EMDiffuse-n demonstrated superior resolution ratio versus baselines, Uncertainty threshold: 0.12 (predictions with uncertainty below 0.12 considered reliable), Acquisition speedup: up to 18× reduction in acquisition time reported (denoising restoring high-quality images from noisy images), LPIPS: EMDiffuse-r outperformed CARE, PSSR, and RCAN across LPIPS, FSIM: EMDiffuse-r outperformed baselines (higher FSIM), Resolution Ratio: EMDiffuse-r outperformed baselines (higher resolution ratio), Fourier Ring Correlation: Indicates EMDiffuse-r captures high-frequency details present in ground truth, Uncertainty threshold: 0.12 (predictions with uncertainty below threshold considered reliable), Acquisition speedup: 36× increase in EM imaging speed (by super-resolving 6 nm pixel to 3 nm pixel), LPIPS: vEMDiffuse-i produced lower LPIPS than 3D-SRU-Net and ITK-cubic interpolation (quantitative improvement reported), FSIM: vEMDiffuse-i produced higher FSIM than 3D-SRU-Net and ITK-cubic interpolation, Resolution Ratio: Improved versus baselines (higher resolution ratio), IoU (organelle segmentation): vEMDiffuse-i generated volume achieved IoU scores similar to ground truth isotropic volume; for some experiments ER and mitochondria IoU > 0.9 (up to axial resolution = 64 nm), Uncertainty: Predictions had uncertainty values below the uncertainty threshold (0.12) indicating reliability, LPIPS/FSIM/Resolution Ratio: vEMDiffuse-a outperformed CARE and ITK-cubic interpolation on LPIPS, FSIM, and resolution ratio in downsampled OpenOrganelle experiments (Supplementary Fig. 23c)., IoU (organelle segmentation): Reported example IoU values in figure captions: 0.60, 0.98, 0.62, 0.94 (these illustrate anisotropic vs vEMDiffuse-a vs isotropic comparisons across datasets/organelle types); vEMDiffuse-a improved IoU and 3D reconstruction fidelity., Uncertainty: Generated volumes had uncertainty values below threshold indicating reliability for MICrONS and FANC demonstrations., IoU: Segmentation models trained on isotropic OpenOrganelle liver volume applied to interpolated, anisotropic, vEMDiffuse-i generated, and ground truth isotropic volumes; vEMDiffuse-i generated volume achieved IoU similar to isotropic volume (Fig. 3e). |
| Application Domains | Electron microscopy (EM) ultrastructural imaging, Volume electron microscopy (vEM) / Connectomics, Cell biology (organelle-level 3D ultrastructural analysis), Large-scale tissue array tomography datasets (e.g., MICrONS, FANC) |
147. MatterSim: A Deep Learning Atomistic Model Across Elements, Temperatures and Pressures, Preprint (May 10, 2024)
| Category | Items |
|---|---|
| Datasets | MatterSim training dataset (~17M structures), MPF-TP, Random-TP, MPF-Alkali-TP, MPF2021, MPtrj (variants: MPTrj-random-1k, MPTrj-highest-stress-1k), Alexandria (and Alexandria-1k, Alexandria-MP-ICSD), PhononDB (MDR phonon database), MatBench (MatBench Discovery and other MatBench tasks), RSS-generated dataset (this work) / Random Structure Search outputs, Experimental Gibbs free energy references (FactSage / SISSO-derived analytical forms) |
| Models | Graph Neural Network, Message Passing Neural Network, Transformer, Multi-Layer Perceptron |
| Tasks | Regression, Binary Classification, Ranking |
| Learning Methods | Supervised Learning, Active Learning, Ensemble Learning, Pre-training, Fine-Tuning, Zero-Shot Learning, Uncertainty Quantification, End-to-End Learning, Knowledge Distillation |
| Performance Highlights | MAE_energy_on_MPF-TP: 36 meV/atom, chemical_accuracy_equivalent: 43 meV/atom, MP_Gap_MAE: 0.1290 eV, logGVRH_MAE: 0.0608 (log GPa), logKVRH_MAE: 0.0488 (log GPa), Dielectric_MAE: 0.2516 (unitless), Phonons_MAE: 26.022 cm^-1, jdft2d_MAE: 32.762 meV/atom, MAE_max_phonon_frequency: 0.87 THz, MAE_average_phonon_frequency: 0.76 THz, MAE_DOS: 0.64 (integrated MAE of DOS; units as in SI), Li2B12H12_max_force_error_reduction: Model reproduces similar accuracy with only 15% of data compared to training from scratch, uncertainty_reduction: Maximum error notably reduced after active learning (no single scalar provided), data_efficiency_gain: finetune-30 (30 configurations) achieves similar performance to scratch-900 (900 configurations) — 97% reduction in data, self_diffusion_D_finetune_corrected: 1.862×10^-5 cm^2/s (error below 20% vs experiment 2.3–2.4×10^-5 cm^2/s), MatBench_Discovery_F1: 0.83, formation_energy_MAE: 0.026 eV/atom (reported ~0.03 eV/atom in Table S2), MD_success_rate_across_families: >90% success rate across tested material families, snapshot_energy_error: <50 meV/atom (mean error of energy lower than 50 meV/atom for example MD snapshots), MAE_vs_PBE_QHA_free_energy_up_to_1000K: <10 meV/atom (sub-10 meV/atom), MAE_vs_experiment_over_200_materials: 15 meV/atom (0–1000K range comparison to experimental in SI), MgO_B1-B2_transition_pressure_at_300K: 584 GPa predicted (compared to experimental 429–562 GPa and first-principles 520 GPa) |
| Application Domains | Materials discovery / computational materials science, Atomistic simulations and molecular dynamics, High-pressure / high-temperature materials (earth mantle conditions, high-pressure synthesis), Inorganic solids (bulk crystals, MOFs, 2D materials, molecular crystals, polymers, interfaces, surfaces), Thermodynamics (Gibbs free energy and phase diagram prediction), Electronic and mechanical property prediction (band gaps, elastic moduli, dielectric constants), Liquid water simulation (ab initio level properties and dynamics), High-throughput screening and random structure search (RSS) |
146. Robotic synthesis decoded through phase diagram mastery, Nature Synthesis (May 2024)
| Category | Items |
|---|---|
| Datasets | 35 quaternary oxides (containing 27 unique elements) |
| Models | None |
| Tasks | Experimental Design, Ranking, Optimization, Data Generation, Decision Making |
| Learning Methods | None |
| Performance Highlights | success_rate: 32/35 (91%), similar_or_greater_purity_count: 31 compounds, unique_successes_by_predicted_precursors: 6 targets, examples_of_metastable_products_and_energies_meV_per_atom: LiNbWO6 (10 meV/atom), LiZnBO3 (8 meV/atom), KTiNbO5 (1 meV/atom), Li3Y2(BO3)3 (39 meV/atom), ranking_criterion: lowest point on the convex hull prioritized; high inverse-hull energy favoured, failure_modes_count: 3 targets not synthesized by either predicted or conventional precursors |
| Application Domains | Materials science, Inorganic oxide synthesis, Battery cathodes and solid-state electrolytes, Automated/high-throughput experimentation, Self-driving labs (closed-loop materials discovery) |
145. Navigating phase diagram complexity to guide robotic inorganic materials synthesis, Nature Synthesis (May 2024)
| Category | Items |
|---|---|
| Datasets | Materials Project phases and formation energies (used to construct convex hulls), ASTRAL robotic laboratory experimental dataset, Text-mined database of traditional solid-state synthesis recipes (Kononova et al.), Candidate reaction list (Supplementary Data 1), Shared experimental data on figshare |
| Models | Graph Neural Network, None (physics-based convex hull / DFT-driven precursor selection — not in provided model list) |
| Tasks | Experimental Design, Optimization, Recommendation, Hyperparameter Optimization |
| Learning Methods | Active Learning, Supervised Learning |
| Performance Highlights | total_experiments: 224, targets_tested: 35, predicted_precursors_success_rate: 32/35 (91%), targets_with_>=20%_higher_phase_purity_using_predicted_precursors: 15, targets_only_synthesized_by_predicted_precursors: 6, targets_with_similar_yields: 16, targets_where_traditional_better: 4, example_reaction_energy_LiBaBO3_overall: -336 meV per atom, LiBO2 + BaO reaction_energy: -192 meV per atom, LiBaBO3_inverse_hull_energy: -153 meV per atom, observed_failure_energy_region: ΔE_reaction > -70 meV per atom; inverse hull energy > -50 meV per atom (region where predicted precursors often failed or were less certain) |
| Application Domains | Inorganic materials synthesis, Solid-state synthesis of multicomponent oxides (oxides, phosphates, borates), Intercalation battery cathode materials, Solid-state electrolytes, Robotic/automated laboratory experimentation |
144. Designing semiconductor materials and devices in the post-Moore era by tackling computational challenges with data-driven strategies, Nature Computational Science (May 2024)
| Category | Items |
|---|---|
| Datasets | Carolina materials database (hypothetical materials databases), Merchant et al. expanded training dataset, Schmidt et al. training dataset (stepwise transfer learning), Public computational materials databases (generic / unspecified), Multi-fidelity datasets (DFT, hybrid functionals, GW, experimental) |
| Models | Generative Adversarial Network, Variational Autoencoder, Diffusion Model, Transformer, Graph Neural Network, Message Passing Neural Network, Convolutional Neural Network, Multi-Layer Perceptron, XGBoost, Support Vector Machine, Naive Bayes, Random Forest, Gaussian Process |
| Tasks | Graph Generation, Data Generation, Regression, Optimization, Feature Selection, Classification, Anomaly Detection, Hyperparameter Optimization |
| Learning Methods | Active Learning, Transfer Learning, Fine-Tuning, Pre-training, Reinforcement Learning, Ensemble Learning, Supervised Learning, Unsupervised Learning |
| Performance Highlights | DFT_optimization_success_rate: 93.5%, convex_hull_threshold_fraction: 75% below 0.1 eV per atom, training_set_size_growth: from 1e4 to ~1e7 (dataset expansion reported), explored_design_space_size: up to 1 billion compounds (as part of stepwise transfer learning), performance_note: progressively enhanced chemical and structural diversity of training set, speedup_BSE: 1 to 2 orders of magnitude (computational efficiency), nanowire_length_applicability: predicted electrical conductivity for systems with lengths up to 17.5 nm |
| Application Domains | Semiconductor materials discovery / design, Crystal structure prediction (CSP), Device fabrication optimization (growth conditions, interface design), Thermal transport prediction and thermal management, Electrical transport prediction (carrier mobility, conductivity), Optical properties and excited-state properties prediction (bandgaps, BSE/GW acceleration), Autonomous / closed-loop experimental synthesis and optimization (perovskite solar cells example), Semiconductor manufacturing testing (wafer defect detection, ML-guided design-for-test, lithography mask optimization), Spintronic materials and superconductors (materials design applications) |
143. Universal chemical programming language for robotic synthesis repeatability, Nature Synthesis (April 2024)
| Category | Items |
|---|---|
| Datasets | χDL files (.xdl) and Chemputer graph files (.json), HPLC / NMR / MS experimental data (spectra, chromatograms, conversions, yields), Experimental result records for specific syntheses (yields, purities for compounds 3,4,5,7,8,9,13, etc.) |
| Models | None |
| Tasks | Optimization, Experimental Design, Data Generation, Automation (framed as Experimental Design / Optimization on platforms) |
| Learning Methods | Online Learning, Active Learning, Transfer Learning |
| Performance Highlights | yield_compound_8_after_workup_optimization: 71%, yield_compound_8_counter-validated: 72%, TIDA_protection_initial_canada: 59%, TIDA_protection_scotland_final_validated: 59% (first attempt at Scotland), later 71% after workup optimization, CDI_amide_coupling_Kinova_conversion: 83% conversion (without final purification), CDI_amide_coupling_Chemputer_complete_conversion: complete conversion by endpoint HPLC, CDI_amide_coupling_Chemputer_Scotland_yield: 93% yield, CDI_amide_coupling_Chemputer_upscaled_benzylamine_yield: 74% yield, compound_3_yield_original: 45%, compound_4_yield_from_3_original: 82%, compound_3_yield_optimized_chemputer: 88%, compound_4_yield_from_3_optimized: 47% |
| Application Domains | Synthetic organic chemistry, Automated chemical synthesis, Laboratory robotics / laboratory automation, Analytical chemistry (online HPLC, NMR, MS integration), Digital chemistry / chemical informatics |
141. Crystal Structure Assignment for Unknown Compounds from X-ray Diffraction Patterns with Deep Learning, Journal of the American Chemical Society (March 27, 2024)
| Category | Items |
|---|---|
| Datasets | ICSD top-100 structure types (simulated XRD patterns), ICSD overall (metadata reported), RRUFF experimental XRD patterns (test set), ICSD top-101−110 structure types (used for OOD test) |
| Models | Convolutional Neural Network, ResNet, Ensemble (union of submodels) |
| Tasks | Multi-class Classification, Classification, Binary Classification, Out-of-Distribution Learning, Dimensionality Reduction, Clustering |
| Learning Methods | Supervised Learning, Unsupervised Learning, Mini-Batch Learning, Backpropagation, Ensemble Learning, Out-of-Distribution Learning |
| Performance Highlights | test_accuracy_on_same_dataset: 99.89%, test_accuracy_on_top-100_structure_types: 80.0%, test_accuracy_with_only_confidence_combination: 65.7% (combinatorial classification using confidence value alone), experimental_accuracy_on_RRUFF: 81.3% (on 80 experimental patterns), OOD_detection_accuracy_at_R0=0.6: 89.1%, in-distribution_confidence_range: 0.9–1.0 (for RCNet tested on same dataset), out-of-distribution_confidence_range: 0–0.1 (for RCNet tested on different subsets), but occasional C>0.9 leading to misclassification, optimized_alpha_test_accuracy: 80.0% at α = 0.7, ablation_effect_of_removing_RCNet#2: accuracy increase of ~17% for α=0; drop to ~4% for α=0.7 |
| Application Domains | Materials science, Inorganic crystallography / crystal structure identification, X-ray diffraction (XRD) pattern analysis, High-throughput experimentation / autonomous (self-driving) laboratories, Mineralogy (experimental validation using RRUFF) |
140. Machine-Learning Assisted Screening Proton Conducting Co/Fe based Oxide for the Air Electrode of Protonic Solid Oxide Cell, Advanced Functional Materials (March 18, 2024)
| Category | Items |
|---|---|
| Datasets | PAA database (proton absorption amount) - 792 samples, 29 features |
| Models | XGBoost, Random Forest, Decision Tree |
| Tasks | Regression |
| Learning Methods | Supervised Learning, Ensemble Learning, Bagging, Boosting |
| Performance Highlights | RMSE: 0.021, MAE: 0.01, R-squared: 0.901, RMSE: 0.022, MAE: 0.012, R-squared: 0.892 |
| Application Domains | Proton-conducting solid oxide cells (P-SOCs), Materials discovery and screening for air electrode materials, Perovskite oxide materials (Co/Fe-based ABO3), Electrochemistry: fuel cells and electrolysis (hydrogen production) |
139. Autonomous reaction Pareto-front mapping with a self-driving catalysis laboratory, Nature Chemical Engineering (March 2024)
| Category | Items |
|---|---|
| Datasets | Fast-Cat in-house experimental dataset (autonomously generated reaction data) |
| Models | Feedforward Neural Network, Gaussian Process |
| Tasks | Regression, Optimization, Hyperparameter Optimization, Feature Selection |
| Learning Methods | Supervised Learning, Ensemble Learning, Active Learning, Batch Learning, Hyperparameter Optimization |
| Performance Highlights | L1_experiments_to_pareto_front: 60, L2-L6_experiments_per_ligand: 40, resource_use_for_L1_campaign: less than 500 ml solvent, 2 mmol total ligand, L1_SN_range_obtained: 0.37–0.85 (l/b 0.8–5.5) |
| Application Domains | Homogeneous catalysis, Hydroformylation of olefins (1-octene), Catalyst and ligand discovery/benchmarking, Flow chemistry (gas–liquid segmented flow reactors), Process development for fine/specialty chemicals, Autonomous experimentation / self-driving laboratories |
137. Digital twins in medicine, Nature Computational Science (March 2024)
| Category | Items |
|---|---|
| Datasets | Contrast-enhanced MRI cohort (Arevalo et al.), MRI time-course datasets for glioblastoma (Swanson and colleagues), Breast tumor imaging dataset (Wu et al.), Study of cardiac sarcoidosis patients (Shade et al.), Hypertrophic cardiomyopathy cohort (O’Hara et al.), Archimedes diabetes model / UVA/Padova type 1 diabetes simulator datasets, The Cancer Genome Atlas (TCGA), Human Cell Atlas, Electronic health record (EHR) datasets and wearable data (general references) |
| Models | Bayesian Network, Multi-Layer Perceptron, Feedforward Neural Network |
| Tasks | Binary Classification, Survival Analysis, Time Series Forecasting, Regression, Feature Extraction, Image Segmentation, Synthetic Data Generation, Clustering, Control |
| Learning Methods | Supervised Learning |
| Performance Highlights | None |
| Application Domains | Cardiology (arrhythmia risk prediction, catheter ablation planning, atrial fibrillation management), Oncology (tumor progression modeling, treatment planning, predictive oncology), Critical care / ICU (sepsis modeling, scoring systems, decision support), Endocrinology (type 1 diabetes closed-loop insulin delivery), Infectious disease (immune-system-focused digital twins, SARS-CoV-2 response), Pharmacology / Drug development (virtual clinical trials, pharmacometrics, virtual populations), Preventive medicine / wearable-based monitoring |
135. A comprehensive transformer-based approach for high-accuracy gas adsorption predictions in metal-organic frameworks, Nature Communications (March 01, 2024)
| Category | Items |
|---|---|
| Datasets | pre-training MOF/COF structures (collected + generated, total >631,000), hMOF_MOFX_DB, CoRE_MOFX_DB, CoRE_MAP_DB (RASPA generated), ToBaCCo.3.0 generated MOFs, Other structure collections referenced (hMOF, CoRE MOF, CCDC, CoRE COF, GCOFs), COF CH4 adsorption dataset (CoRE COFs at 300 K), Other generated property data (Zeo++ outputs) |
| Models | Transformer, Attention Mechanism, Multi-Head Attention, Feedforward Neural Network, Graph Neural Network |
| Tasks | Regression, Ranking, Feature Extraction, Dimensionality Reduction, Clustering |
| Learning Methods | Self-Supervised Learning, Pre-training, Fine-Tuning, Supervised Learning, Transfer Learning, Representation Learning, Multi-Task Learning |
| Performance Highlights | pre-training_representation_accuracy_R2: 0.98, R2_hMOF_MOFX_DB: 0.98, R2_CoRE_MOFX_DB: 0.92, R2_CoRE_MAP_DB: 0.83, R2_Kr_unknown: 0.85, R2_Xe_unknown: 0.41, R2_all_unknown_gases_min: 0.35 (paper states prediction accuracy above 0.35 for all unknown gases), R2_CH4_in_COFs: 0.76, R2_structural_features_hMOF: >0.99, R2_CoRE_MAP_DB_without_pretraining: 0.70, R2_CoRE_MAP_DB_with_pretraining: 0.83 |
| Application Domains | Materials science, Nanoporous materials / metal-organic frameworks (MOFs), Covalent-organic frameworks (COFs), Gas separation and adsorption (industrial gas separation, carbon capture, noble gas purification), High-throughput materials screening / computational materials discovery |
134. Automated synthesis of oxygen-producing catalysts from Martian meteorites by a robotic AI chemist, Nature Synthesis (March 2024)
| Category | Items |
|---|---|
| Datasets | DFT-simulated dataset of high-entropy hydroxides (29,902 compositions), Robot-driven experimental dataset (243 experiments), LIBS spectral dataset (elemental analysis), Search space of candidate compositions |
| Models | Multi-Layer Perceptron, Gaussian Process |
| Tasks | Regression, Optimization, Data Generation |
| Learning Methods | Supervised Learning, Pre-training, Fine-Tuning, Transfer Learning, Backpropagation |
| Performance Highlights | Pearson_r_for_ΔGOH_prediction: 0.998, Pearson_r_for_ΔGO−OH*_prediction: 0.998, Pearson_r_for_Δq_prediction: 0.996, Pearson_r_for_predicted_vs_measured_overpotential: 0.963, train_test_split: 80% train / 20% test, trained_on: 243 experimental datasets (re-training) + simulated descriptors, Model-guided_OPT_η10: 445.1 mV, Improvement_vs_best_experimental: 37.1 mV lower than best pilot experiment (482.2 mV), Exp-guided_OPT_η10: 467.4 mV, Bayesian_iterations: 280 iterations |
| Application Domains | Materials science, Electrocatalysis (oxygen evolution reaction, OER), Automated chemical synthesis / robotics, Planetary science / in-situ resource utilization for Mars, Computational materials discovery (MD, DFT, ML integration) |
133. Data-Driven Design for Metamaterials and Multiscale Systems: A Review, Advanced Materials (February 22, 2024)
| Category | Items |
|---|---|
| Datasets | J-CFID (CFID), OQMD-8 (OQMD), Orthotropic mechanical metamaterials dataset (freeform pixelated unit cells), Large orthotropic dataset (Wang et al.), TPMS dataset, Multiclass lattice dataset, MetaMine / NanoMine data repositories (meta-materials collection), Pixel/voxelated freeform unit cell datasets (examples), Photonic/metasurface datasets (class-based and freeform) |
| Models | Convolutional Neural Network, Multi-Layer Perceptron, Variational Autoencoder, Autoencoder, Generative Adversarial Network, Conditional GAN, Denoising Diffusion Probabilistic Model, Gaussian Process, Decision Tree, Recurrent Neural Network, Graph Neural Network, Encoder-Decoder, Autoencoder -> PCA / Latent-space visualization (Dimensionality Reduction) [model family], Variational Autoencoder-coupled regressor (VAE + regressor), Fourier Neural Operator (related operator learning mention), Gaussian Mixture Models (in density-based inverse methods) |
| Tasks | Regression, Classification, Dimensionality Reduction, Clustering, Image Generation, Synthetic Data Generation, Optimization, Decision Making, Experimental Design, Feature Extraction, Data Augmentation |
| Learning Methods | Supervised Learning, Unsupervised Learning, Semi-Supervised Learning, Reinforcement Learning, Active Learning, Representation Learning, Dimensionality Reduction, Adversarial Training, End-to-End Learning, Fine-Tuning, Supervised + Generative (conditional generation) |
| Performance Highlights | example_training_size: can be effective with small datasets (example: trained on local patches to achieve field-prediction with only 10 design samples in one work), latent_dim_examples: 16D latent representation (Wang et al.) used and visualized via PCA; VAE trained on large datasets (e.g., 240k), qualitative: hPINNs achieved same objective value but produced simpler/smoother solutions with faster convergence (qualitative) compared to PDE-constrained adjoint-based optimization. |
| Application Domains | Metamaterials (general), Mechanical metamaterials, Optical / photonic metasurfaces, Acoustic / phononic metamaterials, Thermal metamaterials / thermal design, Magneto-mechanical metamaterials, Multiscale structural design (engineering: compliant mechanisms, cloaking, morphing, implants), Materials science (crystal datasets, formation energy analysis) |
132. Extracting accurate materials data from research papers with conversational language models and prompt engineering, Nature Communications (February 21, 2024)
| Category | Items |
|---|---|
| Datasets | Bulk modulus test dataset (hand-curated), Critical cooling rate dataset (R_c1 ground truth), Critical cooling rate dataset (R_c2 ChatExtract), Bulk modulus tables/figures subset, Yield strength of high-entropy alloys dataset (ChatExtract outputs) |
| Models | GPT, Transformer |
| Tasks | Classification, Binary Classification, Information Retrieval, Structured Prediction, Text Classification, Named Entity Recognition |
| Learning Methods | Zero-Shot Learning, Prompt Learning, Pre-training, Fine-Tuning |
| Performance Highlights | precision_overall: 90.8%, recall_overall: 87.7%, single-valued_precision: 100%, single-valued_recall: 100%, multi-valued_precision: 100%, multi-valued_recall: 82.7%, precision_overall: 70.1%, recall_overall: 65.4%, single-valued_precision: 100%, single-valued_recall: 88.5%, multi-valued_precision: 97.3%, multi-valued_recall: 55.9%, precision_overall: 61.5%, recall_overall: 62.9%, single-valued_precision: 74.1%, single-valued_recall: 87.7%, multi-valued_precision: 87.3%, multi-valued_recall: 53.5%, precision_overall: 42.7%, recall_overall: 98.9%, single-valued_precision: 100%, single-valued_recall: 100%, multi-valued_precision: 99.2%, multi-valued_recall: 98.4%, precision_overall: 70.0%, recall_overall: 54.7%, single-valued_precision: 100%, single-valued_recall: 76.9%, multi-valued_precision: 86.6%, multi-valued_recall: 45.7%, table_classification_precision: 95%, table_classification_recall: 98%, datapoint_extraction_precision: 91%, datapoint_extraction_recall: 89%, precision: 80%, recall: 82%, standardized_precision: 91.9%, standardized_recall: 84.2%, R_c2_raw_extracted: 634 values, R_c1_ground_truth: 721 entries (raw) |
| Application Domains | Materials science, Automated literature data extraction, Materials property database construction (bulk modulus, critical cooling rates for metallic glasses, yield strength for high-entropy alloys), Scientific NLP (information extraction from papers: text, tables, figure captions) |
130. A generative artificial intelligence framework based on a molecular diffusion model for the design of metal-organic frameworks for carbon capture, Communications Chemistry (February 14, 2024)
| Category | Items |
|---|---|
| Datasets | hMOF dataset (hypothetical MOF dataset), GEOM dataset, OChemDb (Open Chemistry Database), Generated GHP-MOFassemble artifacts (linker and MOF pools produced by this paper), CoRE DB and Cambridge Structural Database (references) |
| Models | Diffusion Model, Graph Neural Network, Graph Neural Network |
| Tasks | Synthetic Data Generation, Graph Generation, Regression, Classification, Feature Extraction, Data Generation |
| Learning Methods | Generative Learning, Pre-training, Supervised Learning, Ensemble Learning |
| Performance Highlights | initial_linker_samples: 64,800 (540 fragments × 20 samples × 6 connection-atom counts), after_hydrogen_addition: 56,257, with_dummy_atoms: 16,162, after_element_filter (remove S, Br, I): 12,305, Model1_R2: 0.932, Model1_MAE: 0.098 m mol g−1, Model1_RMSE: 0.171 m mol g−1, Model2_R2: 0.937, Model2_MAE: 0.100 m mol g−1, Model2_RMSE: 0.170 m mol g−1, Model3_R2: 0.936, Model3_MAE: 0.099 m mol g−1, Model3_RMSE: 0.170 m mol g−1, Ensemble_MAE (reported): 0.093 m mol g−1 (on 10% test set), Ensemble_std_threshold: 0.2 m mol g−1 (approximately 96% of test predictions had std < 0.2), Classification_accuracy (threshold 2 mmol g−1): 98.4% (13,551/13,765 on test set), Balanced_accuracy: 90.7%, training_dataset_for_pretraining: GEOM dataset (~37M conformations / 450k molecules), diversity_indicator: Tanimoto similarity distribution between generated linkers and hMOF linkers: majority around 0.3–0.4 (indicating novelty); internal diversity (IntDiv1/IntDiv2) increased with sampled atoms, screened_structures_input_to_regressor: 18,770 assembled MOFs, predicted_high_performing_by_ensemble: 364 MOFs (predicted > 2 mmol g−1 via ensemble mean+std criterion), downstream_stable_after_MD: 102 MOFs (lattice parameter change <5%), GCMC_confirmed_high_performing: 6 MOFs with CO2 adsorption > 2 mmol g−1 |
| Application Domains | Materials Science, Computational Chemistry, Metal-Organic Framework (MOF) design, Carbon capture / CO2 adsorption, Molecular generative design (method transfer from drug discovery to materials) |
129. Autonomous execution of highly reactive chemical transformations in the Schlenkputer, Nature Chemical Engineering (February 2024)
| Category | Items |
|---|---|
| Datasets | Zenodo repository: .xdl and .json procedure files and full analytical data for each synthesis |
| Models | None |
| Tasks | Control, Planning, Experimental Design, Optimization, Decision Making |
| Learning Methods | None |
| Performance Highlights | None |
| Application Domains | Synthetic chemistry, Organometallic chemistry, Inert-atmosphere (air-/moisture-sensitive) synthesis, Laboratory automation / robotic chemical synthesis, Analytical chemistry (inline NMR and UV–vis), Chemical process safety (automated handling of pyrophoric reagents), Chemical engineering / apparatus design |
128. Automated self-optimization, intensification, and scale-up of photocatalysis in flow, Science (January 26, 2024)
| Category | Items |
|---|---|
| Datasets | RoboChem Datasets (Zenodo, Robochem Datasets v1), Robochem campaign — HAT alkylation campaign dataset, Robochem campaign — Decatungstate-enabled trifluoromethylthiolation datasets, Robochem campaign — Oxytrifluoromethylation campaign dataset, Robochem campaign — Aryl trifluoromethylation datasets, Robochem campaign — C(sp2)–C(sp3) cross-electrophile coupling datasets |
| Models | Gaussian Process, No explicit ML architecture specified (classical surrogate-based Bayesian optimization) |
| Tasks | Optimization, Experimental Design, Data Generation, Hyperparameter Optimization |
| Learning Methods | Model-Based Learning, Active Learning |
| Performance Highlights | yield_percent_HAT_alkylation: >95, isolated_yield_percent_HAT_scaleup: 99, n_experiments_HAT: 19, convergence_in_runs: 9, time_HAT_campaign: 4 hours, productivity_increase_trifluoromethylthiolation: 70-100x (productivity vs reported batch or literature model systems), n_experiments_per_substrate: 18-36, scale_up: 5 mmol (isolated yields closely matched NMR yields), max_space_time_yield_increase_oxytrifluoromethylation: up to 565-fold, n_experiments_per_substrate: 14-25, residence_time_min: as low as 10 s, internal_volume_for_short_residence: 0.26 mL, n_experiments_per_substrate_aryl_trifluoromethylation: 17-35, time_per_campaign: 11-24 hours, observed_improvement: substantial enhancement in yield and productivity vs literature, yield_increase_example_compound_17: from 37% (literature) to 77% (after 60 experiments), n_experiments_compound_17: 60, time_compound_17: 58 hours |
| Application Domains | Photocatalysis, Synthetic organic chemistry, Flow chemistry / continuous-flow photochemistry, Medicinal chemistry (drug-like molecules and functionalization), Agrochemical chemistry, Chemical process intensification and scale-up / process development |
127. A dynamic knowledge graph approach to distributed self-driving laboratories, Nature Communications (January 23, 2024)
| Category | Items |
|---|---|
| Datasets | Aldol condensation closed-loop optimisation dataset (Cambridge & Singapore), Knowledge graph provenance triples (experiment provenance) |
| Models | Gaussian Process |
| Tasks | Optimization, Experimental Design, Data Generation |
| Learning Methods | Evolutionary Learning, Online Learning |
| Performance Highlights | highest_yield_%: 93, number_of_runs: 65, best_environment_factor: 26.17, best_space-time_yield_g_per_L_per_h: 258.175 |
| Application Domains | chemical reaction optimisation, flow chemistry, laboratory automation / self-driving laboratories (SDLs), digital twin / knowledge graph representations for scientific labs, experiment provenance and FAIR data in chemical sciences |
125. Universal machine learning aided synthesis approach of two-dimensional perovskites in a typical laboratory, Nature Communications (January 02, 2024)
| Category | Items |
|---|---|
| Datasets | High-throughput experimental synthesis dataset (training/test), Prediction set from PubChem, Experimental validation set (selected predicted compounds), Literature-reported successful sample (collected as successful data) |
| Models | Support Vector Machine, Decision Tree, Gradient Boosting Tree, Generalized Linear Model |
| Tasks | Binary Classification, Classification, Feature Extraction, Feature Selection, Dimensionality Reduction |
| Learning Methods | Supervised Learning, Ensemble Learning |
| Performance Highlights | AUC: 0.85, Reported accuracy (stated): 85%, Misclassification_note: Only 1 out of 10 molecules of 2D perovskites is misclassified by the ML model (quoted in text), Experimental validation success rate (ML-guided): 61.5% (8 out of 13 predicted compounds successfully synthesized), Chemical intuition success rate (baseline): 16.4% (13/79? reported), Improvement factor: Approximately 4× increase in success rate relative to traditional chemical intuition |
| Application Domains | Materials discovery / materials science, Two-dimensional hybrid organic–inorganic perovskites (2D HOIPs), specifically 2D AgBi iodide perovskites, Experimental synthesis planning and acceleration, Optoelectronic materials (photodetectors, LEDs), X-ray imaging materials / detectors, Chemoinformatics and molecular descriptor-based ML |
124. Active learning guides discovery of a champion four-metal perovskite oxide for oxygen evolution electrocatalysis, Nature Materials (January 2024)
| Category | Items |
|---|---|
| Datasets | Initial 30 resynthesized ABO3 four-metal perovskite oxides, Augmentation set: 10 additional synthesized perovskite oxides, Combined training set (30 initial + 10 augmented), Enumerated candidate set (in silico): 10,101 candidate structures, Mechanism-labelled dataset (AEM vs LOM): curated from literature, Experimental XRD vs simulated/generated XRD pairs |
| Models | Message Passing Neural Network, Graph Neural Network, Feedforward Neural Network / (general) Neural Network (bootstrap ensemble) |
| Tasks | Regression, Binary Classification, Clustering, Synthetic Data Generation, Feature Extraction, Representation Learning |
| Learning Methods | Active Learning, Supervised Learning, Unsupervised Learning, Ensemble Learning, Data Augmentation, Synthetic Data Generation, Representation Learning |
| Performance Highlights | accuracy (5-fold CV, OO-1): 0.882, r.m.s.e. (5-fold CV, OO-1): 0.033, r.m.s.e. (simulated vs experimental XRD positions & intensities): 0.0874, accuracy (5-fold CV, OO-2): 0.897, r.m.s.e. (5-fold CV, OO-2): 0.025, uncertainty criterion coverage: 100% of 10,101 candidates have prediction uncertainty < 0.020 V (OO-2), accuracy (5-fold CV, mechanism classifier): 0.917, AUC (area under ROC): 0.932, perovskite subset accuracy & AUC: 1.0 (both), predicted overpotential for champion (CPCF): 396 mV (predicted), experimental overpotential for champion (CPCF): 391 mV (measured), prediction vs experiment agreement (paired t-test p-value for simulated vs experimental XRD predictions): 0.4805, initial % candidates below uncertainty threshold (OO-1): 2.87%, final % candidates below uncertainty threshold (OO-2): 100% |
| Application Domains | Materials discovery, Electrocatalysis, Oxygen evolution reaction (OER), Perovskite oxides / inorganic functional materials, Catalyst design and optimization, Materials characterization (XRD-driven representations) |
123. Self-driving laboratories to autonomously navigate the protein fitness landscape, Nature Chemical Engineering (January 2024)
| Category | Items |
|---|---|
| Datasets | Cytochrome P450 dataset, GH1 combinatorial sequence space, SAMPLE experimental dataset (GH1 runs), Reproducibility test set: four diverse GH1 enzymes from Streptomyces species |
| Models | Gaussian Process, Gaussian Process |
| Tasks | Binary Classification, Regression, Optimization, Data Generation, Batch Learning |
| Learning Methods | Supervised Learning, Active Learning, Batch Learning |
| Performance Highlights | classification_accuracy: 0.83, Pearson_r: 0.84, measurements_to_find_thermostable_P450s: 26 (on average for UCB positive and Expected UCB in simulation), sample_efficiency_vs_UCB_and_random: 3- to 4-fold fewer samples required compared to standard UCB and random, thermostability_gain: >= 12 °C (agents discovered GH1 sequences at least 12 °C more stable than initial seed sequences), search_fraction: < 2% of full combinatorial landscape searched, observed_batch_effect: slight benefit to running experiments in smaller batches (as reported; evaluated via 10,000 simulated trials) |
| Application Domains | Protein engineering, Synthetic biology, Enzyme engineering (thermostability optimization), Automated/self-driving laboratories, High-throughput experimental design / closed-loop experimentation |
122. Learning skillful medium-range global weather forecasting, Science (December 22, 2023)
| Category | Items |
|---|---|
| Datasets | ERA5, HRES (and HRES-fc0), TIGGE archive (HRES operational tracks), IBTrACS (International Best Track Archive for Climate Stewardship) Version 4, WeatherBench |
| Models | Graph Neural Network, Message Passing Neural Network, Encoder-Decoder, Convolutional Neural Network, Transformer |
| Tasks | Time Series Forecasting, Regression, Binary Classification |
| Learning Methods | Supervised Learning, Backpropagation, Gradient Descent, Mini-Batch Learning, End-to-End Learning |
| Performance Highlights | RMSE skill score on Z500: improvement of around 7 to 14% (GraphCast vs HRES), Percent of targets where GraphCast outperforms HRES: 90.3% of 1380 targets (89.9% significant at P ≤ 0.05), Outperform Pangu-Weather: GraphCast outperformed Pangu-Weather on 99.2% of 252 targets, Model parameters: 36.7 million, Runtime: produces a 10-day forecast in under 1 minute on a single Google Cloud TPU v4 device, RMSE, ACC comparisons vs HRES: GraphCast has better RMSE and ACC skill curves across lead times for Z500 (plots show superior RMSE and ACC), RMSE-optimized blurring analysis: RMSE-optimized blurring applied to GraphCast still has greater skill than analogous blurring applied to HRES on 88.0% of 1380 targets, Extreme heat precision-recall: GraphCast precision-recall curves above HRES for 5- and 10-day lead times; HRES better at 12-hour lead time (no single scalar AUC provided in main text), Tropical cyclone median track error: GraphCast has lower median track error than HRES over 2018-2021; GraphCast significantly better for lead times 18 hours to 4.75 days (bootstrapped 95% CIs reported in Fig. 3A/B), Atmospheric river IVT RMSE: GraphCast improves prediction of IVT compared with HRES, improvement from ~25% at short lead times to ~10% at longer horizons |
| Application Domains | Medium-range global weather forecasting (meteorology), Severe event prediction (tropical cyclone tracking, atmospheric rivers, extreme temperature), Climate-related forecasting and monitoring (implications for climate trends and retraining recency), Potential applications in other geospatial-temporal forecasting problems (climate, ecology, energy, agriculture, human and biological activity) - suggested |
121. Data-driven analysis and prediction of stable phases for high-entropy alloy design, Scientific Reports (December 18, 2023)
| Category | Items |
|---|---|
| Datasets | Collected HEA experimental dataset (raw), Cleaned HEA dataset (after data cleaning/outlier removal), Augmented / balanced HEA dataset (after synthetic data generation), Per-category cleaned counts (Table 1) |
| Models | XGBoost, Random Forest, Support Vector Machine, Multi-Layer Perceptron |
| Tasks | Multi-class Classification, Classification, Feature Selection, Anomaly Detection, Data Augmentation, Hyperparameter Optimization, Feature Extraction |
| Learning Methods | Supervised Learning, Unsupervised Learning, Ensemble Learning, Bagging, Boosting, Backpropagation, Stochastic Gradient Descent |
| Performance Highlights | accuracy_SS_phases_(BCC,FCC,BCC+FCC): 97%, accuracy_with_AM_included_(4-class_set?): ≈94%, accuracy_with_IM_included_(7 categories): ≈89%, accuracy_all_11phases: 86%, impact_of_synthetic_data: improvement <1% (all models), accuracy_SS_phases(BCC,FCC,BCC+FCC): 97%, accuracy_with_AM_included: 93%, accuracy_with_IM_included_(7 categories): ≈89%, accuracy_all_11phases: 85%, n_trees: 100 (model hyperparameter), accuracy_SS_phases(BCC,FCC,BCC+FCC): 95%, accuracy_with_AM_included: 95%, accuracy_with_IM_included_(7 categories): just above 84%, accuracy_all_11phases: 83%, training_epochs: 128, batch_size: 32, learning_rate: 5.0e-4, accuracy_SS_phases(BCC,FCC,BCC+FCC): 94%, accuracy_with_AM_included: ≈94%, accuracy_with_IM_included_(7 categories): just above 84%, accuracy_all_11_phases: 82%, kernel: 6-degree polynomial (c=5.0, γ=1.0 optimized via grid search) |
| Application Domains | Materials Science, Alloy design, High‑entropy alloy (HEA) discovery and phase prediction, Metallurgy, Materials informatics for clean energy / extreme environment applications |
120. Automated classification of big X-ray diffraction data using deep learning models, npj Computational Materials (December 04, 2023)
| Category | Items |
|---|---|
| Datasets | 171k (derived from Inorganic Crystal Structure Database, ICSD), Baseline synthetic dataset, Mixed synthetic dataset, Large synthetic dataset, RRUFF dataset, Materials Project (MP) dataset, Lattice Augmentation dataset, Seven synthetic datasets (peak-shape/noise variants) |
| Models | Convolutional Neural Network, Convolutional Neural Network, Multi-Layer Perceptron, Deep Neural Network |
| Tasks | Multi-class Classification, Multi-class Classification, Multi-class Classification, Synthetic Data Generation, Data Augmentation, Domain Adaptation |
| Learning Methods | Supervised Learning, Domain Adaptation, Data Augmentation, Synthetic Data Generation |
| Performance Highlights | 7-way_accuracy_on_synthetic_baseline: ≈96%, 230-way_accuracy_on_synthetic_baseline: ≈94%, 7-way_accuracy_RRUFF_trained_on_baseline: 12%, 230-way_accuracy_RRUFF_trained_on_baseline: 12%, 7-way_accuracy_RRUFF_trained_on_baseline: 22%, 230-way_accuracy_RRUFF_trained_on_baseline: 17%, 7-way_accuracy_RRUFF_trained_on_mixed: 35%, 230-way_accuracy_RRUFF_trained_on_mixed: 22%, 7-way_accuracy_RRUFF_trained_on_mixed: 64%, 230-way_accuracy_RRUFF_trained_on_mixed: 53%, 7-way_accuracy_RRUFF_trained_on_large: 74%, 230-way_accuracy_RRUFF_trained_on_large: 66%, 7-way_accuracy_RRUFF_trained_on_souped_large: 86%, 230-way_accuracy_RRUFF_trained_on_souped_large: 77%, F1_RRUFF_trained_on_souped_large: 0.753, F1_RRUFF_trained_on_souped_large: 0.568, 7-way_accuracy_MP_trained_on_souped_large: 75%, 230-way_accuracy_MP_trained_on_souped_large: 45%, 7-way_accuracy_MP_trained_on_souped_large: 54%, 230-way_accuracy_MP_trained_on_souped_large: 25%, 7-way_accuracy_MP_trained_on_souped_large: 67%, 230-way_accuracy_MP_trained_on_souped_large: 36%, accuracy_at_80%_size: 88%, accuracy_at_80%_size: 94%, training_accuracy_convergence: ~98% (models converge to ~98% accuracy on synthetic training data), F1_RRUFF_trained_on_souped_large: 0.859 |
| Application Domains | Materials science, Crystallography / X-ray powder diffraction analysis, High-throughput experimental XRD (in-situ experiments), Materials characterization and phase identification, Alloying and dynamic compression experiments (time-resolved / extreme conditions), Automated experimental data analysis and materials discovery |
119. Autonomous chemical research with large language models, Nature (December 2023)
| Category | Items |
|---|---|
| Datasets | Seven-compound synthesis benchmark (web searcher test set), ECL catalogue samples (Model samples), Perera et al. Suzuki reaction dataset (flow dataset), Doyle Buchwald–Hartwig C–N cross-coupling dataset, Large compound SMILES library (used in computational experiments) |
| Models | GPT, Transformer, Attention Mechanism, Multi-Head Attention |
| Tasks | Planning, Information Retrieval, Text Generation, Control, Experimental Design, Optimization, Language Modeling |
| Learning Methods | Prompt Learning, In-Context Learning, Reinforcement Learning, Fine-Tuning, Embedding Learning, Pre-training |
| Performance Highlights | task_score_scale: 1-5 (5=very detailed and chemically accurate; 3=minimum acceptable score), experimental_validation: GC–MS signals matching reference compounds (Suzuki: peak at 9.53 min matching biphenyl; Sonogashira: peak at 12.92 min matching reference), execution_success: Generated code corrected via documentation search and executed successfully on OT-2 and on ECL (HPLC example executed), normalized_advantage: plotted over iterations (improvement over random strategy), normalized_maximum_advantage (NMA): used to compare convergence and maximal performance, iterations: maximum of 20 iterations; corresponds to 5.2% and 6.9% of total search space for first and second datasets respectively, retrieval_success: Documentation sections selected correctly; ECL functions correctly identified; prompt-to-SLL generated code executed at ECL, sample_retrieval: 1,110 Model samples catalog successfully searchable (sample retrieval worked for queries such as ‘Acetonitrile’) |
| Application Domains | Chemistry (organic synthesis and reaction optimization), Laboratory automation / robotics (liquid handling, heater–shaker, plate readers), Cloud laboratory execution (Emerald Cloud Lab), Experimental design and optimization, Scientific information retrieval and documentation grounding |
118. Scaling deep learning for materials discovery, Nature (December 2023)
| Category | Items |
|---|---|
| Datasets | Materials Project (snapshot March 2021), OQMD (Open Quantum Materials Database) (snapshot June 2021), ICSD (Inorganic Crystal Structure Database) (experimental structures), WBM (Wang, Botti and Marques) dataset snapshot (used for comparison), GNoME-discovered dataset (GNoME generated structures and DFT relaxations / ionic relaxation trajectories), AIRSS-generated random-structure test set, AIMD test sets for MLIP robustness (T=400 K and T=1000 K trajectories), M3GNet dataset (used for comparisons) |
| Models | Graph Neural Network, Message Passing Neural Network, Multi-Layer Perceptron, Graph Neural Network, Ensemble (model ensemble) |
| Tasks | Regression, Binary Classification, Clustering |
| Learning Methods | Active Learning, Supervised Learning, Pre-training, Fine-Tuning, Transfer Learning, Ensemble Learning, Representation Learning, Zero-Shot Learning, Out-of-Distribution Learning |
| Performance Highlights | MAE_meV_per_atom_initial_benchmark: 28, MAE_meV_per_atom_improved_network: 21, MAE_meV_per_atom_final_ensemble: 11, MAE_meV_per_atom_compositional_baseline_ref25: 60, MAE_meV_per_atom_compositional_after_AIRSS_filtering: 40, precision_stable_prediction_composition_only_percent: 33, hit_rate_structural_final_percent: greater than 80, hit_rate_compositional_final_percent: 33, initial_hit_rate_structural_percent: less than 6, initial_hit_rate_compositional_percent: less than 3, previous_work_hit_rate_percent: 1, pretrained_MLIP_parameters: 16.24 million, inference_latency_50_atom_system_ms: 14, inference_throughput_ns_per_day_at_2fs: approx. 12, zero_shot_outperforming_state_of_the_art: pretrained GNoME potential outperforms a state-of-the-art NequIP model trained on hundreds of structures (qualitative claim), classification_test_compositions: 623 unseen compositions used for superionic classification experiments, comparative_performance_statement: NequIP trained on M3GNet data performed better than the M3GNet models trained with energies and forces (M3GNet-EF) reported in ref.62 (qualitative/comparative claim) |
| Application Domains | Materials discovery, Solid-state chemistry / inorganic crystal discovery, Battery materials (Li-ion conductors, solid electrolytes), Layered materials for electronics and energy storage, Interatomic potential development for molecular dynamics simulations, High-throughput computational materials screening |
117. An autonomous laboratory for the accelerated synthesis of novel materials, Nature (December 2023)
| Category | Items |
|---|---|
| Datasets | Materials Project (version 2022.10.28), Inorganic Crystal Structure Database (ICSD), SynTERRA / text-mined synthesis literature corpus (24,304 publications), A-Lab experimental dataset (this work), Filtered candidate sets during materials screening |
| Models | Convolutional Neural Network, XGBoost, Multi-Layer Perceptron, Feedforward Neural Network, Ensemble Learning |
| Tasks | Classification, Regression, Recommendation, Optimization, Experimental Design |
| Learning Methods | Supervised Learning, Active Learning, Reinforcement Learning, Representation Learning, Self-Supervised Learning, Ensemble Learning |
| Performance Highlights | ensemble_size: 100, dropout_rate: 50%, training_epochs: 50, training_epochs: 50, targets_improved: 9 targets (6 had zero yield initially), example_yield_increase: approximately 70% increase in yield for CaFe2P2O9, search_space_reduction: up to 80% reduction in number of possible experiments for some targets |
| Application Domains | Materials science / Solid-state inorganic materials synthesis, Autonomous experimentation / Robotics-integrated laboratory automation, Computational materials (DFT-driven screening and phase stability analysis), Text-mining / NLP for synthesis knowledge extraction, X-ray diffraction analysis and automated crystallographic refinement |
115. Vision-controlled jetting for composite systems and robots, Nature (November 2023)
| Category | Items |
|---|---|
| Datasets | BodyParts3D |
| Models | None |
| Tasks | Depth Estimation, Control, Experimental Design |
| Learning Methods | None |
| Performance Highlights | depth_map_resolution: 64 µm × 32 µm × 8 µm, raw_data_rate: 2 GB s^-1, depth_map_size: 54-megapixel depth map, depth_map_processing_time: < 1 s (to convert image data to 54-megapixel depth map), heightmap_pixels_per_map: 2.36 × 10^9 pixels computed into height map within 2.5 s, scanner_camera_rate: each camera captures 6,000 laser line images per second; four cameras used; each camera captures 9,000 images per scan, printing_throughput: up to 33 ml min^-1, speedup_vs_prior_work: 660 times faster than previous work (MultiFab ref. 33), voxel_throughput: 24 × 10^9 voxels h^-1, voxel_size_used_in_examples: 32 µm × 64 µm × 20 µm (example voxel size), total_assigned_voxels_capacity: 6.15 × 10^11 individually assigned voxels, build_volume: 500 mm × 245 mm × 200 mm, print_speed_z_max: up to 16 mm h^-1 (z-direction), print_head_specs: print heads native resolution 400 DPI; eject droplets ≈ 70 pl at 15 kHz; each inkjet unit contains four print heads (Fujifilm Dimatix SG1024-L), heightmap_generation_rate: height maps computed from 2.36 × 10^9 pixels within 2.5 s; geometrically calibrated to 32 µm × 64 µm × 20 µm pixel resolution, walking_robot_speed: approximately 0.01 m s^-1 (approx. 0.1 body length s^-1), walking_robot_turn_speed: (20/15) ° s^-1 (reported turning speed), pneumatic_joint_pressure_capacity: each joint supports actuation pressures up to 35 kPa, pump_flow_rate: up to 2.3 l min^-1 at 90 beats min^-1, robot_actuation_channels: walker uses four pneumatic networks; valves command pressures between 0 kPa to 250 kPa; valve flow per channel up to 380 l min^-1 |
| Application Domains | Soft Robotics, Additive Manufacturing / 3D Printing, Materials Science (polymer chemistry), Robotics (bioinspired robot systems), Biomedical / Anatomical modeling (hand geometry), Metamaterials, Industrial manufacturing / prototyping |
114. AI-driven robotic chemist for autonomous synthesis of organic molecules, Science Advances (November 2023)
| Category | Items |
|---|---|
| Datasets | ReaxysDB (subset used for training), Synbot experimental dataset for target molecules M1, M2, M3 |
| Models | Transformer, Message Passing Neural Network, Graph Neural Network |
| Tasks | Sequence-to-Sequence, Optimization, Regression |
| Learning Methods | Supervised Learning, Active Learning |
| Performance Highlights | top1_prediction_accuracy_improvement: 4.5-7.0%, M1_reference_conversion_on_Synbot: 86.5%, M1_reported_isolation_yield_reference: 37.7%, M1_target_conversion_set: 91.5%, M1_best_found_conversion: 100.0% (found within first trial of searchspace), M2_reference_conversion_on_Synbot: 15.0%, M2_target_conversion_set: 70.0%, M2_best_found_conversion: 100.0% (at 36th and 37th tryouts), M3_modified_reference_conversion_on_Synbot: 50.9%, M3_target_conversion_set: 80.0%, M3_best_found_conversion: target achieved at 42nd trial, system_throughput: 12 reactions per 24 hours (on average), efficiency_gain_vs_human: at least 6x (Synbot vs human performing two experiments/day), reproducibility_dispensing_MAE: ≤0.73 mg, reproducibility_dispensing_CV: ≤2.55%, conversion_yield_CV_overall: <5%, conversion_yield_CV_late_stage: <2.5%, trials_fraction_of_search_space_to_goal: <1% of trials from total search space |
| Application Domains | organic chemistry / synthetic chemistry, automated laboratory robotics / robotic chemistry, chemical process optimization, materials discovery (functional organic materials, organic electronics, pharmaceuticals) |
113. Machine learning-enabled constrained multi-objective design of architected materials, Nature Communications (October 19, 2023)
| Category | Items |
|---|---|
| Datasets | Unlabeled porosity matrix dataset (~18,000 samples), Labeled FEM simulation dataset (initial labeled set: 95; total labeled set size not explicitly stated), Experimental compression test replicates (Ti6Al4V and pure Zn scaffolds), Repository: GAD-MALL code and datasets |
| Models | Autoencoder, Convolutional Neural Network, Variational Autoencoder, Gaussian Mixture Model, Finite Element Method |
| Tasks | Regression, Multi-objective Optimization, Data Generation, Feature Extraction, Optimization, Image Generation |
| Learning Methods | Unsupervised Learning, Supervised Learning, Active Learning, Representation Learning, Ensemble Learning |
| Performance Highlights | R2: >0.92 (on test dataset), MAE: low (exact value not specified in main text; referenced in Supplementary Fig. 4), latent_dimension: 8, reconstruction_loss: chosen trade-off (variational variant had higher reconstruction loss and was not used), E2500_Y_increase: >30% (yield strength improvement found at rounds 3 and 5), Macro_implant_load-bearing_increase: ≈20% higher experimental load-bearing capacity (ML-inspired vs uniform), Zn_Y_increase: ~20% increase in Y for ML-designed Zn scaffolds (experimental), simulation-experiment_error: <10% (FEM calibrated to experiments), model_selection_metric: average negative log-likelihood vs number of clusters (supplementary analysis) |
| Application Domains | Architected materials / metamaterials, Orthopedic implants / bone scaffold design, 3D printing / Additive manufacturing (laser powder bed fusion), Tissue engineering / biomedical implants, Computational materials design (ML + FEM integration) |
111. Scalable Diffusion for Materials Generation, International Conference on Learning Representations (October 13, 2023)
| Category | Items |
|---|---|
| Datasets | Perov-5, Carbon-24, MP-20, Materials Project (MP) 2021, GNoME (GNoME dataset / structure search database), AIRSS runs / AIRSS-converged compositions, UniMat training set (all experimentally verified stable materials + additional found via search/substitution) |
| Models | Denoising Diffusion Probabilistic Model, U-Net, Convolutional Neural Network, Attention Mechanism, Variational Autoencoder, Generative Adversarial Network, Graph Neural Network, PointNet, Transformer |
| Tasks | Data Generation, Graph Generation, Synthetic Data Generation |
| Learning Methods | Generative Learning, Supervised Learning, Self-Supervised Learning |
| Performance Highlights | Proxy_validity_MP-20%: 97.2, Proxy_coverage_MP-20%: 89.4, Proxy_recall_MP-20%: 99.8, Proxy_precision_MP-20%: 99.7, ∆Ef_vs_CDV_AE (eV/atom): -0.216, EfReductionRate_vs_CDV_AE: 0.863, Num_Stable_vs_MP2021: UniMat 414 vs CDV AE 56 (Ed<0 wrt MP 2021), Num_Metastable_vs_MP2021: UniMat 2157 vs CDV AE 90 (Ed<25 meV/atom wrt MP 2021), Num_Stable_vs_GNoME: UniMat 32 vs CDV AE 1 (Ed<0 wrt GNoME convex hull), AIRSS_convergence_rate: 0.55 (AIRSS baseline), UniMat_convergence_rate: 0.81 (overall; computed 0.817 in Appendix C), ∆Ef_vs_AIRSS (eV/atom): -0.68, EfReductionRate_vs_AIRSS: 0.80, Ablation_model_size_small_structural_validity_%: 95.7, Ablation_model_size_small_composition_validity_%: 86.0, Ablation_model_size_small_recall_%: 99.8, Ablation_model_size_small_precision_%: 99.3, Ablation_model_size_large_structural_validity_%: 97.2, Ablation_model_size_large_composition_validity_%: 89.4, Ablation_model_size_large_recall_%: 99.8, Ablation_model_size_large_precision_%: 99.7, Proxy_validity_Perov-5%: 100 (CDV AE Perov-5), Proxy_COV_Perov-5%: 98.5, ∆Ef_vs_MP-20test (CDV AE, MP-20 test): 0.279, EfReductionRate_vs_MP-20_test: 0.083, Num_Stable_MP2021: 56 stable (Ed<0), Num_Metastable_MP2021: 90 metastable (Ed<25 meV/atom), Proxy_metrics_reference: Included as a baseline in proxy metric comparisons (Table 1) — specific values reported per dataset in Table 1, Example_Perov-5_validity%: 100 (LM Perov-5 reported 100 in Table 1), MP-20validity%: 95.8 (LM on MP-20 in Table 1) |
| Application Domains | Materials discovery, Computational materials science, Crystal structure generation, Accelerating materials synthesis and screening (in silico), High-throughput DFT verification workflows |
110. Universal machine learning for the response of atomistic systems to external fields, Nature Communications (October 12, 2023)
| Category | Items |
|---|---|
| Datasets | H2O toy dataset (single equilibrium geometry), N-methylacetamide (NMA) dataset, Liquid water dataset (full), Liquid water exemplary dataset for FieldSchNet comparison (special dataset) |
| Models | Message Passing Neural Network, Multi-Layer Perceptron, Graph Neural Network |
| Tasks | Regression |
| Learning Methods | Supervised Learning, Multi-Task Learning, Supervised Learning, Mini-Batch Learning, Backpropagation, Representation Learning, End-to-End Learning |
| Performance Highlights | NMA_energy_RMSE_eV: 0.0053, NMA_dipole_RMSE_Debye: 0.028, NMA_polarizability_RMSE_a.u.: 0.51, LiquidWater_FIREANN-wF_forces_RMSE_meV_per_Angstrom: 39.4, 200-configs_FIREANN_forces_RMSE_meV_per_Angstrom: 54.5, 200-configs_FIREANN_polarizability_RMSE_a.u.: 2.1, 200-configs_FieldSchNet_forces_RMSE_meV_per_Angstrom: 245.4, 200-configs_FieldSchNet_polarizability_RMSE_a.u.: 165.1, full_dataset_FIREANN_forces_RMSE_meV_per_Angstrom: 45.5, full_dataset_FIREANN_polarizability_RMSE_a.u.: 2.5, full_dataset_FieldSchNet_forces_RMSE_meV_per_Angstrom: 184.7, full_dataset_FieldSchNet_polarizability_RMSE_a.u.: 12.9, training_time_per_epoch_A100_80GB_FIREANN_minutes: 2.4, training_time_per_epoch_A100_80GB_FieldSchNet_minutes: 7.6, H2O_toy_energy_vs_DFT_behavior: qualitatively exact reproduction of rotational and field-intensity dependence (no numeric RMSE reported) |
| Application Domains | computational chemistry / quantum chemistry, molecular spectroscopy (IR and Raman), molecular dynamics and path-integral MD (nuclear quantum effects), condensed-phase / liquid water simulations, materials and periodic systems under external electric fields, electrochemistry (potential application), plasmonic chemistry (potential application), tip-induced catalytic reactions (potential application) |
109. In Pursuit of the Exceptional: Research Directions for Machine Learning in Chemical and Materials Science, Journal of the American Chemical Society (October 11, 2023)
| Category | Items |
|---|---|
| Datasets | Inorganic Crystal Structure Database (ICSD), Materials Project, AFLOW, OQMD, SuperCon database, Pearson’s Crystal Dataset, Experimental hardness measurements dataset, Training set used by Pogue et al. / Meredig/Stanev studies |
| Models | Random Forest, Gradient Boosting Tree, Autoencoder, Transformer, Generative Adversarial Network, Convolutional Neural Network |
| Tasks | Regression, Classification, Optimization, Multi-objective Optimization, Active Learning, Anomaly Detection, Clustering, Sequence-to-Sequence, Data Generation, Representation Learning |
| Learning Methods | Supervised Learning, Unsupervised Learning, Active Learning, Transfer Learning, Multi-Task Learning, Ensemble Learning, Evolutionary Learning, Reinforcement Learning, Representation Learning, Feature Selection |
| Performance Highlights | percent_above_superhard_threshold_at_0.5N: 0.1%, percent_above_superhard_threshold_at_5N: 0.01%, example_predicted_Hv: Sc2OsB6 ≈ 38 GPa, relative_experiments_to_find_high_Tc: ≈ 1/3 of experiments required by random search |
| Application Domains | Materials science (inorganic materials, superconductors, superhard materials, high-entropy alloys), Chemistry (organic synthesis, retrosynthesis, reaction prediction), Experimental automation / self-driving laboratories, Catalysis and nanoparticle synthesis, Photovoltaics and perovskite solar cells, Mechanical property prediction and design (hardness, elastic moduli), Microscopy and spectroscopy (automated characterization and anomaly detection), Drug discovery and biomedical polymers (mentioned as areas ML contributes) |
108. Finite-difference time-domain methods, Nature Reviews Methods Primers (October 05, 2023)
| Category | Items |
|---|---|
| Datasets | Ground-truth FDTD simulation data (coarse-mesh and dense-mesh results referenced in ref. 99 and other refs), Coarse-mesh FDTD simulation outputs (used as input features) and dense-mesh FDTD results (ground-truth), FDTD outputs used for PINN / physics-based deep learning (examples in refs. 97, 98) |
| Models | Convolutional Neural Network, Long Short-Term Memory, Multi-Layer Perceptron |
| Tasks | Regression, Time Series Forecasting, Sequence-to-Sequence |
| Learning Methods | Supervised Learning, Self-Supervised Learning, Monte Carlo Learning |
| Performance Highlights | qualitative: rapidly and accurately computed scattering parameters; achieved results ‘in a fraction of the simulated time needed for a dense grid FDTD simulation’, qualitative: replicating the FDTD solution with excellent accuracy (qualitative statement; no numeric error metrics reported in text) |
| Application Domains | Computational electromagnetics (full-wave Maxwell simulations), Microwave circuits and devices (S-parameter analysis), Photonics and optics (photonic crystals, metasurfaces, metalenses), Plasmonics and nanophotonics, Biomedical imaging and biosensing (OCT, PWS, SERS, MRI coil design, dosimetry), Geophysics and ionospheric propagation (ground-penetrating radar, Earth–ionosphere waveguide), Metamaterials (negative-index lenses, cloaks, leaky-wave antennas), Quantum electromagnetics / quantum photonics (quantum FDTD, Hong–Ou–Mandel modelling), Multiphysics coupling (electromagnetic–thermal, electromagnetic–circuit, quantum–electromagnetic) |
107. Inverse design of chiral functional films by a robotic AI-guided system, Nature Communications (October 04, 2023)
| Category | Items |
|---|---|
| Datasets | Experimental dataset of 1493 chiral films, Parameter space specification (prescreened combinations) |
| Models | Multi-Layer Perceptron, Feedforward Neural Network, Generative Adversarial Network |
| Tasks | Regression, Clustering, Optimization, Data Generation, Hyperparameter Optimization, Data Augmentation |
| Learning Methods | Supervised Learning, Unsupervised Learning, Adversarial Training, Stochastic Gradient Descent, Hyperparameter Optimization |
| Performance Highlights | MAE: 0.04, RMSE: 0.06, R2: 0.985, MAE: 0.09, RMSE: 0.15, R2: 0.95, Experimental_g_abs_max: 1.9, Experimental_g_lum_max: 1.9 |
| Application Domains | Chiral materials / chiroptical materials, Photonics / optical materials, Materials science (film fabrication and characterization), Display technology (multiplex laser display / color filters), Luminescent materials (circularly polarized luminescence with perovskite quantum dots), Automated experimentation / robotic chemistry |
106. Retrosynthesis prediction with an interpretable deep-learning framework based on molecular assembly tasks, Nature Communications (October 03, 2023)
| Category | Items |
|---|---|
| Datasets | USPTO-50K, USPTO-FULL, USPTO-MIT, Nine Tanimoto similarity-based USPTO-50K splits, Leaving Group (LG) vocabulary / database, Multi-step pathway planning cases (provided by authors) |
| Models | Transformer, Graph Neural Network, Graph Attention Network, Graph Convolutional Network, Message Passing Neural Network, Multi-Layer Perceptron, Feedforward Neural Network, Long Short-Term Memory, Radial Basis Function Network, Seq2Seq |
| Tasks | Sequence-to-Sequence, Graph Generation, Link Prediction, Node Classification, Multi-class Classification, Multi-task Learning, Planning |
| Learning Methods | Multi-Task Learning, Contrastive Learning, Pre-training, Self-Supervised Learning, Representation Learning, Dynamic Learning Rate Scheduling (polynomial decay with warm-up) |
| Performance Highlights | USPTO-50K (reaction class known) top-1: 66.8%, USPTO-50K (reaction class known) top-3: 88.0%, USPTO-50K (reaction class known) top-5: 92.5%, USPTO-50K (reaction class known) top-10: 95.8%, USPTO-50K (reaction class unknown) top-1: 57.7%, USPTO-50K (reaction class unknown) top-3: 79.2%, USPTO-50K (reaction class unknown) top-5: 84.8%, USPTO-50K (reaction class unknown) top-10: 91.4%, USPTO-FULL top-1: 51.4%, USPTO-FULL top-3: 70.7%, USPTO-FULL top-5: 74.7%, USPTO-FULL top-10: 79.2%, USPTO-MIT top-1: 60.3%, USPTO-MIT top-3: 81.6%, USPTO-MIT top-5: 86.4%, USPTO-MIT top-10: 90.5%, Multi-step pathway planning literature correspondence: 86.9% of single-step reactions (153/176) correspond to reported reactions via SciFinder search, LGM accuracy (reaction-type given vs unknown range reported): increases from 65.4% to 73.2% when reaction type label is provided (text reports improvement range), RCP accuracy (reaction-type given vs unknown range reported): RCP accuracy increased from 81.6% to 91.2% when reaction type label is provided (text reports improvement range), multi-step planning: pathways identified: 101 pathways, single-step matches to literature: 153/176 single steps matched (~86.9%) via SciFinder search, re-ranking improvements: significant improvement in top-1, top-3, top-5, top-10 accuracies when re-ranking predictions from RetroXpert, GLN, and NeuralSym (box-plot based results reported; exact numeric deltas not provided in main text) |
| Application Domains | Organic chemistry, Computer-assisted synthetic planning (retrosynthesis), Drug development / medicinal chemistry, Computational chemistry / cheminformatics |
105. Accelerating science with human-aware artificial intelligence, Nature Human Behaviour (October 2023)
| Category | Items |
|---|---|
| Datasets | Inorganic materials corpus (Tshitoyan et al. dataset), MEDLINE (biomedical literature), DrugBank candidate drug pool, Comparative Toxicogenomics Database (CTD) curated drug–disease associations, ClinicalTrials.gov (COVID-19 trials), Candidate inorganic compounds pool, Author disambiguation resources |
| Models | Graph Neural Network, Graph Convolutional Network, GraphSAGE, Autoencoder |
| Tasks | Link Prediction, Ranking, Recommendation, Node Classification, Feature Extraction |
| Learning Methods | Unsupervised Learning, Representation Learning, Feature Learning |
| Performance Highlights | thermoelectricity_precision_full_graph: 62%, ferroelectricity_precision_full_graph: 58%, photovoltaics_precision_full_graph: 74%, thermoelectricity_precision_authorless_graph: 48%, ferroelectricity_precision_authorless_graph: 50%, photovoltaics_precision_authorless_graph: 58%, discoverer_top50_precision_thermoelectric_and_ferroelectric_1yr: 40%, discoverer_top50_precision_photovoltaics_1yr: 20% |
| Application Domains | Materials science (inorganic materials, thermoelectricity, ferroelectricity, photovoltaics), Drug discovery and drug repurposing, Biomedical research (disease–drug associations), COVID-19 therapeutics and vaccines, Science of science / bibliometrics (prediction of scientific discoveries and discoverers) |
104. A foundation model for generalizable disease detection from retinal images, Nature (October 2023)
| Category | Items |
|---|---|
| Datasets | MEH-MIDAS (Moorfields Diabetic imAge dataSet), Kaggle EyePACS, Reference 34 OCT dataset (SPECTRALIS/related), MEH-AlzEye, UK Biobank, Kaggle APTOS-2019, IDRiD, MESSIDOR-2, PAPILA, Glaucoma Fundus, JSIEC, Retina (Kaggle cataractdataset), OCTID |
| Models | Vision Transformer, Transformer, Autoencoder, Multi-Layer Perceptron, ResNet, Generalized Linear Model |
| Tasks | Image Classification, Multi-class Classification, Binary Classification, Time Series Forecasting, Survival Analysis, Feature Extraction |
| Learning Methods | Self-Supervised Learning, Supervised Learning, Transfer Learning, Fine-Tuning, Contrastive Learning, Generative Learning, Pre-training |
| Performance Highlights | AUROC_APTOS-2019: 0.943 (95% CI 0.941, 0.944), AUROC_IDRID: 0.822 (95% CI 0.815, 0.829), AUROC_MESSIDOR-2: 0.884 (95% CI 0.88, 0.887), AUROC_CFP: 0.862 (95% CI 0.86, 0.865), AUROC_OCT: 0.799 (95% CI 0.796, 0.802), AUROC_MyocardialInfarction_CFP: 0.737 (95% CI 0.731, 0.743), sensitivity_MyocardialInfarction_CFP: 0.70, specificity_MyocardialInfarction_CFP: 0.67, AUROC_HeartFailure: 0.794 (95% CI 0.792, 0.797), AUROC_IschaemicStroke: 0.754 (95% CI 0.752, 0.756), AUROC_Parkinsons: 0.669 (95% CI 0.65, 0.688), AUROC_wetAMD_DINO: 0.866 (95% CI 0.864, 0.869), AUROC_ischaemicStroke_DINO: 0.728 (95% CI 0.725, 0.731), AUROC_wetAMD_CFP_SL-ImageNet: 0.83 (95% CI 0.825, 0.836), AUROC_age_logistic_regression: 0.63 |
| Application Domains | Ophthalmology (retinal imaging), Medical imaging / Clinical diagnosis, Oculomics (using retinal images to predict systemic diseases), Cardiovascular disease risk prediction, Neurodegenerative disease prediction (e.g., Parkinson’s), Clinical prognostics (wet-AMD conversion) |
103. Accelerated discovery of multi-elemental reverse water-gas shift catalysts using extrapolative machine learning approach, Nature Communications (September 21, 2023)
| Category | Items |
|---|---|
| Datasets | Initial catalyst dataset (Iteration = 0), Closed-loop experimental dataset (final), Catalyst candidate composition grid |
| Models | Random Forest, Multi-Layer Perceptron |
| Tasks | Regression, Optimization, Clustering, Feature Selection, Feature Extraction, Experimental Design, Hyperparameter Optimization |
| Learning Methods | Supervised Learning, Ensemble Learning, Representation Learning, Feature Learning, Active Learning, Batch Learning |
| Performance Highlights | R^2 (cross-validation, final / best achieved): 0.81, number_of_tested_candidates_experimentally: 300 (255 ML-predicted), number_of_discovered_superior_catalysts: >100, discovery_outcome: identified Pt(3)/Rb(1)-Ba(1)-Mo(0.6)-Nb(0.2)/TiO2 as optimal catalyst; discovered >100 catalysts outperforming prior best, R^2 (explorative model with 8-dim elemental descriptor representation): achieved highest prediction accuracy among the three representation strategies at initial stage; overall best R^2 = 0.81 reported after iterations |
| Application Domains | heterogeneous catalysis, catalyst discovery / materials discovery, materials informatics, chemical reaction engineering (reverse water-gas shift, CO2 conversion), experimental design / autonomous discovery workflows |
102. Accurate proteome-wide missense variant effect prediction with AlphaMissense, Science (September 19, 2023)
| Category | Items |
|---|---|
| Datasets | ClinVar, ProteinGym (MAVE collection), Additional MAVE benchmark (this study), Deciphering Developmental Disorders (DDD) de novo variants, Proteome-wide possible missense variants (all possible single amino acid substitutions), Primate variant population data (and human population frequency data), gnomAD (used implicitly for observed/absent variants and allele frequency strata), UK Biobank / GeneBass summary statistics (complex trait associations) |
| Models | Transformer, Autoencoder, Attention Mechanism, Self-Attention Network |
| Tasks | Binary Classification, Classification, Regression, Language Modeling, Representation Learning, Ranking |
| Learning Methods | Self-Supervised Learning, Pre-training, Fine-Tuning, Weakly Supervised Learning, Knowledge Distillation, Ensemble Learning, Representation Learning, Fine-Tuning |
| Performance Highlights | ClinVar_auROC: 0.94, ClinVar_per_gene_auROC_average: 0.95, Proteome_predictions_total: 71 million missense variant predictions (saturating human proteome), Classified_fraction_at_90pct_precison_on_ClinVar: 92.9% resolved vs EVE 67.1%, Proteome_fraction_likely_pathogenic: 32% (22.8 million), Proteome_fraction_likely_benign: 57% (40.9 million), ProteinGym_mean_Spearman: 0.514, Additional_MAVE_mean_Spearman: 0.45, Subset_25_human_proteins_mean_Spearman_all_methods_comparison: 0.474, Example_SHOC2_Spearman: 0.47, Example_GCK_Spearman: 0.53, auROC_underpowered_genes_cell_essentiality: 0.88, LOEUF_auROC_underpowered: 0.81, auROC_rest_of_proteome_alphaMissense: 0.8, auROC_rest_of_proteome_LOEUF: 0.82, ESM1v_MAVE_example_Spearman: 0.459, ESM1v_ClinVar_auROC: reported in comparisons (lower than AlphaMissense; exact number not given in main text for ClinVar) |
| Application Domains | Human genetics / clinical variant interpretation, Molecular biology / protein function and mutational scanning, Rare disease diagnostics, Complex trait genetics / population genomics, Computational biology / proteome-scale variant effect prediction, Functional genomics (MAVE integration), Drug target and biomedical research prioritization |
101. Demonstration of an AI-driven workflow for autonomous high-resolution scanning microscopy, Nature Communications (September 07, 2023)
| Category | Items |
|---|---|
| Datasets | Few-layer WSe2 dark-field experimental dataset (APS live experiment), Pre-acquired dark-field WSe2 image (numerical validation dataset), Cameraman (standard image) - training image, MIT/USC-SIPI/scikit-image images (for simulations) |
| Models | Multi-Layer Perceptron, Radial Basis Function Network, Gaussian Process |
| Tasks | Image Inpainting, Experimental Design, Optimization, Feature Extraction |
| Learning Methods | Supervised Learning, Pre-training, Batch Learning, Transfer Learning |
| Performance Highlights | stable_reconstruction_coverage: 27%, experimental_reconstruction_sufficient_coverage: <25%, experiment_time_saving: ≈65% (~80 minutes saved for the 200x40 demonstration), decision_making_overhead: ≤2% (computation ≈0.15 s to compute new positions per iteration), decision_time_per_iteration: ≈0.15 s (compute new positions) ; ≈42 s to scan batch of 50 positions ; total ≈0.37 s to process diffraction patterns and communicate measurements (per iteration overhead numbers reported), FAST_candidate_calc_time_comparison: FAST: ≈1.5 s (200×40 image on low-power CPU) vs GP: ≈6 s (50×50 image on NVIDIA DGX-2) reported from literature comparison, reconstruction_quality_metrics: NRMSE and SSIM curves show FAST achieves lower NRMSE and higher SSIM at substantially lower sampling percentages compared to raster grid (RG), uniform random (UR), and low-discrepancy (LDR) sampling; exact numeric NRMSE/SSIM values not tabulated in text, final_reconstruction_method: Biharmonic inpainting applied to measured points to generate final images (higher-quality reconstructions shown), sparse_sampling_effectiveness: FAST reproduces flake boundaries and bubbles with high fidelity at much lower measurement percentages than static sampling methods (qualitative and NRMSE/SSIM improvements shown); reconstruction stabilized between 15-20% in experimental demonstration (visual convergence by ≈20%) |
| Application Domains | Scanning microscopy, X-ray microscopy (dark-field scanning diffraction microscopy), Synchrotron beamline experiments (Advanced Photon Source), Materials characterization (2D materials, WSe2 thin films), Autonomous / self-driving laboratory experimentation |
100. A principal odor map unifies diverse tasks in olfactory perception, Science (September 2023)
| Category | Items |
|---|---|
| Datasets | GS-LF (Good Scents + Leffingwell databases), Prospective validation panel dataset (this study), QC subset for GC-MS/GC-O analyses, Compiled likely-odorants list (~500k), Dravnieks Atlas of Odor Character Profiles, DREAM Olfaction Prediction Consortium dataset (Keller et al.), Abraham et al. detection-threshold dataset, Snitz et al. perceptual similarity dataset |
| Models | Graph Neural Network, Random Forest, Support Vector Machine, Linear Model |
| Tasks | Multi-label Classification, Regression, Dimensionality Reduction, Feature Extraction, Representation Learning, Out-of-Distribution Learning |
| Learning Methods | Supervised Learning, Representation Learning, Transfer Learning, Fine-Tuning, Mini-Batch Learning, Ensemble Learning |
| Performance Highlights | cross_validation_AUROC: 0.89, prospective_eval_better_than_median_panelist_percent: 53%, example_per-molecule_correlation_RGNN: 0.63 (example shown), per-label_surpassed_median_panelist: 30/55 labels (55%), statistical_comparison_to_prior_state_of_art: paired two-tailed Student’s t-test p = 3.3e-7 (GNN vs previous SOTA on same data), prospective_eval_better_than_median_panelist_percent: 41%, example_per-molecule_correlation_RRF: 0.45 (example shown), triplet_discordance_correct_percent: 19% (baseline model correctly predicts empirical discordance only 19% of time), descriptor_applicability_vs_SVM: POM-linear model outperforms chemoinformatic SVM baseline on Dravnieks, DREAM (Keller), and current data (higher target correlation R), detection_threshold_vs_SVM: POM-linear model outperforms chemoinformatic SVM baseline on Abraham et al. detection-threshold data (higher correlation R), perceptual_similarity_vs_SVM: POM-linear model outperforms chemoinformatic SVM baseline on Snitz et al. perceptual similarity data (higher correlation R), triplet_discordance_prediction_percent: 50% (GNN model correctly predicted counterintuitive structure-odor discordance in 50% of triplets), chemical_class_sulfur_R: R = 0.52 (mean correlation for sulfur-containing molecules) |
| Application Domains | olfaction / smell perception, cheminformatics / chemical space mapping, sensory neuroscience, psychophysics, flavor and fragrance industry |
99. Learning heterogeneous reaction kinetics from X-ray videos pixel by pixel, Nature (September 2023)
| Category | Items |
|---|---|
| Datasets | In situ STXM images of carbon-coated LiFePO4 nanoparticles, Auger Electron Microscopy (AEM) carbon-coating intensity maps, Ptychography images (ex situ) |
| Models | State Space Model, Gaussian Process |
| Tasks | Regression, Structured Prediction, Image-to-Image Translation, Uncertainty Quantification |
| Learning Methods | Maximum A Posteriori, Model-Based Learning |
| Performance Highlights | training_RMSE: 6.8%, validation_RMSE: 9.6 ± 0.9%, experimental_noise_estimate_sigma_e: ≈0.07 (7%), training_RMSE_at_rho2=0.01: 6.0%, training_RMSE_at_rho2->∞: 10.6%, pixel-to-pixel_correlation_js_vs_AEM: -0.4 (correlation coefficient), training_RMSE: 6.8%, validation_RMSE: 9.6 ± 0.9% |
| Application Domains | battery materials / Li-ion battery electrodes, operando microscopy / in situ imaging, energy materials (electrochemistry), materials science (phase-separating solids), non-destructive imaging and characterization |
98. Material symmetry recognition and property prediction accomplished by crystal capsule representation, Nature Communications (August 25, 2023)
| Category | Items |
|---|---|
| Datasets | Materials Project (MP) dataset, MatBench dataset |
| Models | Capsule Network, Transformer, Multi-Layer Perceptron, Long Short-Term Memory, DenseNet, Convolutional Neural Network, Graph Neural Network, Graph Convolutional Network, Graph Attention Network, Attention Mechanism |
| Tasks | Regression, Feature Extraction, Representation Learning, Dimensionality Reduction, Clustering |
| Learning Methods | Supervised Learning, Maximum Likelihood Estimation, Variational Inference, Attention Mechanism, Representation Learning, Feature Extraction, End-to-End Learning |
| Performance Highlights | MAE_bandgap_MatBench: 0.181 eV, MAE_formation_energy_MatBench: 0.0161 eV/atom, MAE_bandgap_test_6027: 0.25 eV, MAE_formation_energy_test_6027: 0.0184 eV/atom, MAE_bandgap_test_baseline_MLP: 0.58 eV, MAE_bandgap_test_baseline_DenseNet: 0.55 eV, MAE_bandgap_test_baseline_TFN: 0.49 eV, MAE_bandgap_test_baseline_SE(3)_transformer: 0.86 eV, MAE_reduction_vs_CGCNN_bandgap: 39.1%, MAE_reduction_vs_CGCNN_formation_energy: 52.6%, MAE_reduction_vs_MEgNet_bandgap: 6.2%, MAE_reduction_vs_MEgNet_formation_energy: 36.1%, MAE_reduction_vs_SchNet_bandgap: 23.3%, MAE_reduction_vs_SchNet_formation_energy: 26.1%, MAE_bandgap_test_baseline_EGNN: 0.76 eV |
| Application Domains | Materials science / computational materials discovery, Crystal property prediction (bandgap, formation energy), Solid-state physics / electronic structure prediction, Machine learning for scientific interpretation (symmetry recognition and representation learning) |
97. ChatGPT Chemistry Assistant for Text Mining and the Prediction of MOF Synthesis, Journal of the American Chemical Society (August 16, 2023)
| Category | Items |
|---|---|
| Datasets | MOF synthesis dataset (text-mined), Embedding dataset of text segments, Held-out test set for ML prediction |
| Models | GPT, Random Forest, Representation Learning |
| Tasks | Named Entity Recognition, Text Summarization, Binary Classification, Information Retrieval, Sequence Labeling, Feature Selection, Dimensionality Reduction, Text Generation, Question Answering, Clustering |
| Learning Methods | Prompt Learning, Few-Shot Learning, Zero-Shot Learning, Supervised Learning, Representation Learning, Ensemble Learning, Pre-training, Embedding Learning |
| Performance Highlights | precision: >95% (aggregate across processes), recall: >90% (aggregate across processes), F1: >92% (aggregate across processes), per-parameter precision/recall/F1: 90-99% reported in abstract for ChemPrompt-guided extraction, accuracy: 87% (average on held-out set), F1: 92% (on held-out set), other_metrics_reported: Precision, Recall, Area Under the Curve were evaluated (values not all explicitly listed; F1/accuracy quoted), embedding_dimensionality: 1536, speed_improvement: 28−37% faster on subsequent readings when storing embeddings locally (reduces processing time by 15−20s per paper described), qualitative_performance: Effective at filtering irrelevant content but can miss synthesis segments that mix characterization or crystallographic data (mitigated by passing mid-relevance to classifier) |
| Application Domains | chemistry (metal-organic frameworks synthesis), literature text mining / scientific literature curation, materials science (MOF synthesis outcome prediction), scientific data-to-dialogue systems (chatbot for MOF synthesis Q&A) |
96. Enhancing corrosion-resistant alloy design through natural language processing and deep learning, Science Advances (August 11, 2023)
| Category | Items |
|---|---|
| Datasets | Electrochemical metrics for corrosion-resistant alloys (adapted for pitting potential) |
| Models | Recurrent Neural Network, Long Short-Term Memory, Multi-Layer Perceptron, Feedforward Neural Network, Autoencoder |
| Tasks | Regression, Feature Extraction, Feature Selection, Optimization, Out-of-Distribution Learning |
| Learning Methods | Supervised Learning, Representation Learning, Feature Extraction, Gradient Descent, Backpropagation, Representation Learning |
| Performance Highlights | validation_loss (MAE approx): 150 mV, R2 (average over sixfold cross-validation, test predictions): 0.78 ± 0.06, R2 (previous simple DNN for comparison): 0.61 ± 0.04, validation_loss (MAE approx): 168 mV, R2 (average over sixfold cross-validation, test predictions): 0.66, validation_loss (MAE approx): 170 mV, R2 (average over sixfold cross-validation, test predictions): 0.61 ± 0.04, optimization_learning_rate: 0.0001, method: Multidimensional gradient descent using AugNet to compute derivatives, example_word_effect: introduction of the word ‘potentiostatic’ changed predicted pitting potential from ~177.53 mV to 233.18 mV (error values reported in Table 2) |
| Application Domains | Materials science, Corrosion science / Electrochemistry, Alloy design and optimization, Computational metallurgy, Materials informatics / Machine learning for materials |
95. Applied machine learning as a driver for polymeric biomaterials design, Nature Communications (August 10, 2023)
| Category | Items |
|---|---|
| Datasets | PolyInfo, Khazana, Polymers: a Property Database, Polymer Property Predictor and Database (PPPDB), MatWeb, Block Copolymer Phase Behavior Database (BCDB), Electron Affinity and Ionization Potential Data, Huan et al. polymer dataset (Sci Data 2016), Community Resource for Innovation in Polymer Technology (CRIPT), Simulated datasets (DFT/molecular dynamics), High-throughput experimental datasets (automated copolymer synthesis examples), Property-specific datasets summarized in Table 3 (collection of studies) |
| Models | Random Forest, Gaussian Process, Recurrent Neural Network, Long Short-Term Memory, Multi-Layer Perceptron, Support Vector Machine, Gradient Boosting Tree, LightGBM, Message Passing Neural Network, Variational Autoencoder, Generative Adversarial Network, Linear Model, Graph Neural Network, Gaussian Mixture Model, Random Forest |
| Tasks | Regression, Graph Generation, Optimization, Ranking, Clustering, Dimensionality Reduction |
| Learning Methods | Supervised Learning, Unsupervised Learning, Transfer Learning, Fine-Tuning, Active Learning, Ensemble Learning, Evolutionary Learning |
| Performance Highlights | None |
| Application Domains | Polymeric biomaterials design, Medical devices (catheters, coatings, implants), Drug delivery and polymer excipients, Regenerative medicine (tissue engineering scaffolds), Antifouling coatings & polymers, Biosensors and biologic sensing, 3D printing of tissue engineering scaffolds, Protein/ribonucleoprotein delivery (gene editing), Medical imaging (e.g., 19F MRI agents), Polymer electronics / implantable electronics |
94. Machine Learning Descriptors for Data-Driven Catalysis Study, Advanced Science (August 04, 2023)
| Category | Items |
|---|---|
| Datasets | Additive library (Guo et al.), High-throughput catalyst screening dataset (Nguyen et al.), High-throughput OCM catalyst dataset (Ishioka et al.), Permuted three-element catalyst space (Ishioka et al.), Collected OER experimental dataset (Hong et al.), Materials Project perovskite database screening, DFT-computed perovskites for GPR (Li et al. / related), DFT adsorption datasets for *CO and *OH (Li et al.), HEA structures dataset (molecular dynamics + DFT), Spectral adsorption configurations (Wang et al.), Single-atom-alloy screening dataset (SISSO-based high-throughput), Small experimental ORR dataset (Karim et al.), Robotic experimental overpotential dataset |
| Models | Decision Tree, Random Forest, XGBoost, Linear Model, Gradient Boosting Tree, Support Vector Machine, Gaussian Process, Multi-Layer Perceptron, Graph Convolutional Network, Random Forest (as used for extra-trees / ETR surrogate) |
| Tasks | Regression, Classification, Clustering, Feature Selection, Feature Extraction, Ranking, Hyperparameter Optimization, Dimensionality Reduction |
| Learning Methods | Supervised Learning, Unsupervised Learning, Transfer Learning, Ensemble Learning, Boosting, Representation Learning |
| Performance Highlights | cross_validation_score_range: 0.67-0.84, cross_validation_score_range: 0.67-0.84, Pearson_r_Eads_CO_on_Ag(111): 0.961, Pearson_r_Δe_CO_on_Ag(111): 0.954, RMSE_Eads: 0.015 eV, RMSE_Δe: 0.005 e-, MAE: 0.55 eV, R2: 0.90, MAE: 0.51 eV, R2: 0.84, RMSE_prediction_vs_DFT: ≈0.2 eV, testing_MAE: 0.09 eV, testing_RMSE: 0.12 eV, approximate_scope: GPR used to extend predictions to ≈4000 double perovskites from ~250 DFT calculations, MAE: 0.51 eV, R2: 0.84, Pearson_r_predicted_vs_experimental_overpotentials: 0.878, spectral_ETR_prediction_quality: high (Pearson r typically >0.8; many >0.9 for transfer to diverse systems) |
| Application Domains | Heterogeneous catalysis, Electrocatalysis (CO2 reduction reaction, OER, ORR, NRR, HER), Oxidative coupling of methane (OCM), High-throughput experimental catalyst screening, High-entropy alloy (HEA) catalyst discovery, Single-atom alloy catalysts (SAACs), Perovskite oxide catalysts, Metal-zeolite catalysts, Surface–adsorbate interaction prediction (computational spectroscopy), Catalyst design combining DFT and experimental data |
93. Machine-learning-assisted material discovery of oxygen-rich highly porous carbon active materials for aqueous supercapacitors, Nature Communications (August 01, 2023)
| Category | Items |
|---|---|
| Datasets | Literature-collected dataset for ANN training (Supplementary Table 1), Experimental dataset from this work (hyperporous carbons synthesized and measured in this study) |
| Models | Multi-Layer Perceptron |
| Tasks | Regression |
| Learning Methods | Supervised Learning, Backpropagation |
| Performance Highlights | dataset_size: 288 data points, train_RMSE: 25.0, validation_RMSE: 34.5, test_RMSE: 38.5, validation_MSE_at_best_epoch: 1189 (best training performance at epoch 35), train_RMSE_after_adding_experimental_data: 33.0, validation_RMSE_after_adding_experimental_data: 35.6, test_RMSE_after_adding_experimental_data: 46.2, predicted_max_specific_capacitance_initial (in 1 M H2SO4): 611 F/g, predicted_optimal_features_initial: Smicro = 1502 m2/g, Smeso = 687 m2/g, O content = 20 at.%, N content = 0.5 at.%, predicted_max_specific_capacitance_from_prior literature (comparison): 570 F/g (reference prediction for N/O-doped carbons in 6 M KOH from earlier ANN), predicted_optimal_features_after_reinforcement: Smicro = 1710 m2/g, Smeso = 1050 m2/g, N-doping = 2.3 at.%, O-doping = 20 at.% |
| Application Domains | Materials discovery, Electrochemical energy storage, Supercapacitors (carbon-based electrodes), Materials informatics / data-driven materials design |
92. Scientific discovery in the age of artificial intelligence, Nature (August 2023)
| Category | Items |
|---|---|
| Datasets | 1.6 million organic-light-emitting-diode material candidates, 11 billion synthon-based ligand candidates, ATOM3D, Open Reaction Database, Open Catalyst 2020 (OC20), GuacaMol, Therapeutics Data Commons, 250 million protein sequences (protein sequence corpus), LHC Olympics 2020 (anomaly detection challenge / HEP dataset), Pretrained black-hole waveform models (gravitational-wave waveform datasets) |
| Models | Autoencoder, Variational Autoencoder, Generative Adversarial Network, Normalizing Flow, Diffusion Model, Convolutional Neural Network, Transformer, Graph Neural Network, Graph Convolutional Network, Graph Attention Network, Message Passing Neural Network, Recurrent Neural Network, Boltzmann Machine, Attention Mechanism, Multi-Layer Perceptron |
| Tasks | Anomaly Detection, Image Super-Resolution, Image Denoising, Language Modeling, Structured Prediction, Graph Generation, Regression, Image Generation, Decision Making, Clustering, Sequence-to-Sequence, Ranking |
| Learning Methods | Unsupervised Learning, Self-Supervised Learning, Supervised Learning, Reinforcement Learning, Active Learning, Weakly Supervised Learning, Transfer Learning, Fine-Tuning, Contrastive Learning, Pre-training, Adversarial Training |
| Performance Highlights | speed_improvement: up to six orders of magnitude faster than traditional methods, protein_structure_accuracy: atomic accuracy (described qualitatively as ‘with atomic accuracy, even for proteins whose structure is unlike any of the proteins in the training dataset’) |
| Application Domains | Physics (high-energy physics, particle collisions, astrophysics, gravitational-wave astronomy), Chemistry (drug discovery, small-molecule design, synthesis planning, catalysis), Materials science (materials discovery, organic light-emitting-diode candidates, catalysts), Biology and bioinformatics (protein folding, protein design, genomics, single-cell analysis), Earth sciences (seismology, Earth system science), Medical imaging and healthcare (pathology slides, chest X-rays, MRI, diagnostic applications), Robotics / experimental automation (self-driving labs, autonomous experimentation), Quantum physics and quantum experiments, Computational fluid dynamics and differential-equation-based modelling |
91. Fatigue database of complex metallic alloys, Scientific Data (July 12, 2023)
| Category | Items |
|---|---|
| Datasets | FatigueData-CMA2022 |
| Models | ResNet, Convolutional Neural Network, GPT, BERT, Linear Model |
| Tasks | Image Classification, Semantic Segmentation, Text Classification, Structured Prediction, Regression, Dimensionality Reduction, Clustering, Information Retrieval |
| Learning Methods | Few-Shot Learning, Pre-training, Fine-Tuning, Supervised Learning |
| Performance Highlights | precision: 86%, recall: 94%, F1: 90%, precision: 81%, recall: 90%, F1: 86%, F1: 88%, table data extraction precision: 62%, table data extraction recall: 76%, table data extraction F1: 68%, data extraction precision: 82%, data extraction recall: 51%, data extraction F1: 63%, text data extraction precision: 81%, text data extraction recall: 97%, text data extraction F1: 88% |
| Application Domains | Materials science, Fatigue of complex metallic alloys, Metallic glasses (MGs), Multi-principal element alloys / High-entropy alloys (MPEAs/HEAs), Data-driven materials design and property analysis, Scientific literature mining / automated data curation |
90. Encoding physics to learn reaction–diffusion processes, Nature Machine Intelligence (July 2023)
| Category | Items |
|---|---|
| Datasets | Synthetic 2D Gray–Scott (GS) reaction–diffusion dataset (training measurements), Synthetic 3D Gray–Scott (GS) reaction–diffusion dataset (training measurements), Synthetic 2D / 3D FitzHugh–Nagumo (FN) reaction–diffusion datasets, Synthetic 2D λ–Ω reaction–diffusion dataset, Coefficient identification datasets for 2D Gray–Scott (GS) (S1 and S2), Synthetic Burgers’ equation dataset (used for interpretability example), Additional examples referenced (Kolmogorov turbulent flows, etc.) |
| Models | Convolutional Neural Network, Recurrent Neural Network, Long Short-Term Memory, Multi-Layer Perceptron, ResNet, Feedforward Neural Network, Graph Neural Network, Transformer |
| Tasks | Time Series Forecasting, Image Super-Resolution, Regression, Structured Prediction, Feature Extraction |
| Learning Methods | Supervised Learning, Gradient Descent, Backpropagation, Fine-Tuning, Transfer Learning |
| Performance Highlights | accumulative_RMSE: significantly lower than baselines across considered time-marching interval (exact curves shown in Fig. 2 and Fig. 4), mean_absolute_relative_error_coefficients_inverse: 0.6% (noise-free), 1.61% (10% Gaussian noise) for coefficient identification, parameter_efficiency: PeRCNN uses the least number of trainable parameters among compared models (reported in Supplementary Note E.5), accumulative_RMSE: higher than PeRCNN; performs fairly well on 2D cases but deviates considerably for 3D cases (see Fig. 2 and Fig. 4), accumulative_RMSE: higher than PeRCNN; particularly deviates in 3D cases (see Fig. 2), inverse_identification_error: PeRCNN shows superiority to the PINN (details in Supplementary Table 4), reconstruction: recurrent ResNet unable to reconstruct fine-resolution snapshots in 2D GS case under limited noisy training data (see Supplementary Note E.5.1), training_time_per_epoch: For 3D case, elapsed time for training one epoch by PeRCNN is comparable to that of the ResNet (Supplementary Note E.5), discovery_metrics: precision, recall and relative ℓ2 error of the coefficient vector reported in Supplementary Note F (no single scalar in main text); method recovers governing PDEs completely for clean or mildly noisy data and uncovers majority of terms at 10% noise, example_identified_coefficients: ut = 2.001 × 10−5 Δu − 1.003 uv^2 − 0.04008 u + 0.04008; vt = 5.042 × 10−6 Δv + 1.009 uv^2 − 0.1007 v (from Fig. 6 / main text) |
| Application Domains | Chemistry (reaction systems), Biology (pattern formation, cell proliferation), Geology, Physics (spatiotemporal PDE systems), Ecology (population dynamics), Fluid dynamics (Burgers’, Navier–Stokes references), Epidemiology (mentioned as systems with unknown PDEs), Climate science, Materials science (super-resolution, homogenization), Computational engineering / scientific computing |
88. Using a physics-informed neural network and fault zone acoustic monitoring to predict lab earthquakes, Nature Communications (June 21, 2023)
| Category | Items |
|---|---|
| Datasets | p5270, p5271 |
| Models | Multi-Layer Perceptron |
| Tasks | Regression, Time Series Forecasting |
| Learning Methods | Supervised Learning, Backpropagation, Mini-Batch Learning, Transfer Learning, Pre-training, Fine-Tuning |
| Performance Highlights | test_R2_shear: test R2 > 0.9 for training data >= 20%, test_R2_slip: R2 between 0.75 and 0.87 (varies with training size), RMSE_table: {‘70-10-20’: 0.0923, ‘60-10-20’: 0.0964, ‘50-10-20’: 0.0986, ‘40-10-20’: 0.0948, ‘30-10-20’: 0.108, ‘20-10-20’: 0.145, ‘10-10-20’: 0.148, ‘5-10-20’: 0.1835}, RMSE_table_PINN#1: {‘70-10-20’: 0.092, ‘60-10-20’: 0.0943, ‘50-10-20’: 0.0967, ‘40-10-20’: 0.0938, ‘30-10-20’: 0.1031, ‘20-10-20’: 0.1269, ‘10-10-20’: 0.1336, ‘5-10-20’: 0.1654}, RMSE_table_PINN#2: {‘70-10-20’: 0.09, ‘60-10-20’: 0.0863, ‘50-10-20’: 0.088, ‘40-10-20’: 0.0909, ‘30-10-20’: 0.103, ‘20-10-20’: 0.122, ‘10-10-20’: 0.1227, ‘5-10-20’: 0.1487}, relative_improvement_low_data: PINN models outperform purely data-driven models by roughly 10-15% when training data are scarce (<=20%), test_R2_behavior: For training data >= 20% PINN test R2 > 0.9 for shear stress; PINNs show improved stability (lower variance) and better slip rate prediction, especially PINN #2, general: All transfer-learned (TL) models outperform standalone (trained from scratch) p5271 models across data splits; TL models converge faster., specific_observations: TL PINN #1 consistently outperforms other models across splits after tuning; TL PINN #2 and TL data-driven show similar performance except at 10% where TL PINN #2 significantly outperforms TL data-driven; TL PINN models outperform standalone by large margin when training data scarce (10%)., RMSE_reference: Supplementary Table S2 corroborates TL models have smaller RMSE than standalone; TL PINN #1 has smallest errors per split (numerical RMSEs reported in supplement)., learned_constants_PINN#1_error_range: 2% to 14% deviation from known experimental values across varying training dataset sizes (σ, k, v_l), learned_constants_PINN#2_error_range: 1% to 8% deviation from known experimental values across varying training dataset sizes (σ, K, v_l, ρ); A_intact true value not available, contextual_note: Errors generally increase as training set size decreases |
| Application Domains | Geophysics / Earthquake physics (laboratory earthquake prediction), Seismic monitoring and prediction, Structural health monitoring / nondestructive evaluation (generalizable context), Geothermal reservoir monitoring, induced seismicity risk assessment, CO2 storage and unconventional reservoir monitoring (application relevance discussed) |
87. Discovery of senolytics using machine learning, Nature Communications (June 10, 2023)
| Category | Items |
|---|---|
| Datasets | Training dataset (assembled by authors), Screening libraries (assembled by authors for computational screen), Top predicted hits selected for experimental validation, Experimental validation datasets (cellular assays), Panel of known senolytics used as positives (training) |
| Models | Random Forest, Support Vector Machine, XGBoost, Naive Bayes, Generalized Linear Model, Decision Tree, Message Passing Neural Network |
| Tasks | Binary Classification, Classification, Clustering, Community Detection, Feature Selection, Dimensionality Reduction, Data Augmentation, Feature Extraction |
| Learning Methods | Supervised Learning, Ensemble Learning, Boosting |
| Performance Highlights | precision_5-fold_CV: 0.7 ± 0.16, selected_hits_fraction_screened: 21 / 4340 (0.4%), hit_confirmation_rate_experimental: 3 / 21 = 14.28%, prediction_cutoff: selected compounds with P > 44% (prediction probability), qualitative: high precision, low recall (few false positives but many false negatives), feature_selection: used average reduction of Gini index to select 165 features from 200, qualitative: higher recall but lower precision compared to RF (opposite trade-off to RF), qualitative: substantially outperformed by XGBoost on this dataset (no numeric metrics provided), qualitative: worse performance than SVM and RF in comparisons (Supplementary Table 2) |
| Application Domains | Drug discovery / early-stage virtual screening, Senolytics discovery (targeting cellular senescence), Cheminformatics / computational chemistry, Phenotypic (target-agnostic) screening, Experimental cell biology validation (in vitro senescence models) |
86. Precursor recommendation for inorganic synthesis by machine learning materials similarity from scientific literature, Science Advances (June 09, 2023)
| Category | Items | |||
|---|---|---|---|---|
| Datasets | Text-mined inorganic solid-states synthesis recipes (Kononova et al. dataset and derived expansions used in this work) | |||
| Models | Multi-Layer Perceptron, Feedforward Neural Network, Attention Mechanism, Transformer, Rawcomposition, Magpie encoding, FastText, Multi-Layer Perceptron |
|||
| Tasks | Recommendation, Multi-label Classification, Regression, Feature Extraction, Representation Learning |
|||
| Learning Methods | Self-Supervised Learning, Multi-Task Learning, Representation Learning, Supervised Learning, Backpropagation, Stochastic Learning |
|||
| Performance Highlights | success_rate_within_5_attempts: 82%, second_attempt_success_rate: 73%, first_guess_most_common_baseline_success_rate: 36%, example_predicted_probabilities_LaAlO3: see table: e.g., P(use La2O3 |
cond La2O3)=0.75, P(use Al2O3 | cond Al2O3)=0.73, P(use Al(NO3)3 | cond Al(NO3)3)=0.65, success_rate_within_5_attempts: 68%, success_rate_within_5_attempts: 56%, num_test_materials_applicable: 1985 (out of 2654) due to vocabulary issues, success_rate_within_5_attempts: 66%, success_rate_within_5_attempts: 58% |
| Application Domains | Inorganic materials synthesis, Solid-state synthesis of inorganic materials, Materials science (synthesis planning and recommendation), Autonomous laboratories and recommendation engines for experimental design |
85. The rise of self-driving labs in chemical and materials sciences, Nature Synthesis (June 2023)
| Category | Items |
|---|---|
| Datasets | Open Reaction Database (ORD), Chiral metal halide perovskite nanoparticle experiments, Photocatalyst formulation campaign (hydrogen evolution), Quantum dot / semiconductor nanoparticle synthesis datasets, 3D-printed geometry experiments for mechanical optimization, General datasets generated by self-driving labs (SDLs) |
| Models | Multi-Layer Perceptron, Feedforward Neural Network, Graph Neural Network |
| Tasks | Optimization, Experimental Design, Regression, Image Classification, Clustering, Hyperparameter Optimization |
| Learning Methods | Active Learning, Evolutionary Learning, Supervised Learning, Online Learning, Transfer Learning, Representation Learning |
| Performance Highlights | discovery_speedup: >1,000× faster (referenced for autonomous synthesis–property mapping and on-demand synthesis of semiconductor and metal nanoparticles), notes: specific performance numbers vary by study, photocatalyst_activity: 6× more active than prior art, experiments: 688 experiments in 8-day continuous unattended operation, experiment_count_reduction: 60× fewer experiments than conventional grid search (three-dimensional-printed geometry case), general_benefit: reduced total cost of computation and experimentation when leveraging prior data/models (qualitative), example_reference: transfer learning used in designing lattices for impact protection (ref. 82) |
| Application Domains | Chemical synthesis (organic synthesis, retrosynthesis), Materials science (nanomaterials, thin films, perovskites), Clean energy technologies (photocatalysts, solar materials), Pharmaceuticals / active pharmaceutical ingredients (APIs), Additive manufacturing / mechanical design (3D-printed geometries), Catalysis, Device manufacturing and co-design (materials + device integration) |
84. Combinatorial synthesis for AI-driven materials discovery, Nature Synthesis (June 2023)
| Category | Items |
|---|---|
| Datasets | Inkjet-printing composition libraries (Gregoire et al. implementation), Microfluidic CsPbX nanocrystal parametric maps (droplet-based microfluidic platform), Autonomous solution-based synthesis platforms datasets (Ada, RAPID, MAOSIC), Combinatorial sputter-deposition thin-film libraries, Web of Science publication counts (combinatorial / autonomous materials) |
| Models | None explicitly named from provided model list |
| Tasks | Optimization, Experimental Design, Data Generation, Anomaly Detection, Information Retrieval, Structured Prediction, Regression |
| Learning Methods | Active Learning, Supervised Learning, Generative Learning, Representation Learning, Ensemble Learning |
| Performance Highlights | materials_per_human_intervention: on the order of 10–10^3 materials (for autonomous solution-based platforms), latency: good/low latency for autonomous solution-based workflows, throughput_example: one-to-several unique composition libraries per day for sputter deposition; ~400,000 materials per day achievable for inkjet printing workflows, closed_loop_discovery_examples: demonstrated in refs; qualitative acceleration of discovery reported, latency/automation: good latency and improved decision-making in closed-loop implementations, synthesis_duration: as low as 3–5 seconds per synthesis, in-line_characterization: real-time PL and absorbance used to infer composition/size/distribution, quality_scores: microfluidic nanoparticle synthesis earned ‘good’ for purity and monitoring in the paper’s metric table |
| Application Domains | materials discovery, thin-film materials science, electrocatalysis (oxygen evolution catalysts), photoelectrochemical materials / photoanodes, nanoparticle / nanocrystal synthesis (perovskite nanocrystals), autonomous experimental workflows / self-driving laboratories, data science / literature mining for materials synthesis |
83. Discovering small-molecule senolytics with deep neural networks, Nature Aging (June 2023)
| Category | Items |
|---|---|
| Datasets | Screened compounds (training set), Broad Institute Drug Repurposing Hub (scored subset), Extended Broad Institute library (scored subset), Predicted chemical space (final prediction set), Curated compounds for experimental validation, Validated hits, Protein structural dataset for molecular docking |
| Models | Graph Neural Network, Message Passing Neural Network, Feedforward Neural Network, Random Forest, Ensemble (of Chemprop models) |
| Tasks | Binary Classification, Classification, Regression, Dimensionality Reduction, Ranking |
| Learning Methods | Supervised Learning, Ensemble Learning, Representation Learning, Bayesian Hyperparameter Optimization, Pre-training |
| Performance Highlights | auPRC: 0.243, auPRC_95%_CI: 0.138–0.339, baseline_auPRC: 0.019, prediction_score_range: 2.1e-6 to 0.70 (on 804,959 compounds), working_hit_rate_positive_predictive_value: 11.6% (25 true positives out of 216 high-ranking curated compounds), validated_true_positives_curated: 25/216 (high-ranking), validated_true_negatives_curated: 50/50 (low-ranking negative controls), auPRC_max_observed: 0.15, number_with_PS_gt_0.4_Drug_Repurposing_Hub: 28 (and 284 with PS > 0.1), number_with_PS_gt_0.5_extended_library: 681, number_with_PS_gt_0.4_extended_library: 2,537 |
| Application Domains | aging / geroscience, drug discovery / small-molecule screening, cheminformatics / virtual screening, computational biology / molecular docking, pharmacology / senotherapeutics |
82. A robotic platform for the synthesis of colloidal nanocrystals, Nature Synthesis (June 2023)
| Category | Items |
|---|---|
| Datasets | Gold nanocrystals experimental database, Double-perovskite (Cs2AgIn1−xBixCl6) experimental database, Literature-mined synthesis-parameter dataset for gold NCs, Solvent screening dataset (double-perovskite), Surfactant screening dataset (double-perovskite), RGB color dataset (gold and double-perovskite NCs) |
| Models | SISSO (sure independence screening and sparsifying operator) |
| Tasks | Regression, Experimental Design, Feature Extraction, Data Generation, Optimization |
| Learning Methods | Supervised Learning |
| Performance Highlights | R^2: 0.95, R^2: 0.94, R^2: 0.90, achieved_AR_mean: 4.06, achieved_AR_std: 0.41, achieved_sizes_nm: 78 (nanosized), 749 (microsized) |
| Application Domains | Colloidal nanocrystal synthesis, Materials chemistry / inorganic materials, Perovskite nanocrystals, Robotic/automated chemical synthesis, High-throughput experimentation, Materials discovery and inverse design |
81. Data-driven design of new chiral carboxylic acid for construction of indoles with C-central and C–N axial chirality via cobalt catalysis, Nature Communications (May 31, 2023)
| Category | Items |
|---|---|
| Datasets | rxn1 dataset (Cp*Co(III)/CCA-catalyzed asymmetric C–H alkylation of indoles with central chirality), rxn2 delta dataset (target atroposelective C–H alkylation with axial chirality), virtual screening candidate CCA set, reaction encoding / descriptor set |
| Models | Support Vector Machine, Random Forest, XGBoost |
| Tasks | Regression, Ranking, Optimization, Feature Extraction |
| Learning Methods | Supervised Learning, Transfer Learning, Few-Shot Learning |
| Performance Highlights | Pearson R (10-fold CV, rxn1): 0.859, MAE (10-fold CV, rxn1): 0.179 kcal/mol, Pearson R (direct application to rxn2, base model without transfer): 0.451, MAE (rxn2 base model before delta correction): 0.210 kcal/mol, MAE (rxn2 after delta learning correction): 0.095 kcal/mol, Predicted enantioselectivity (CCA-3, CCA-4, CCA-5): predicted ~89% (CCA-3, CCA-4), 88% (CCA-5), Experimental enantioselectivity (CCA-4): 94% e.e., Prediction error (other tested CCAs CCA-6 to CCA-9): maximum error of 14% e.e., Relative performance: LSVR reported as best in 10-fold CV (Pearson R 0.859, MAE 0.179 kcal/mol); detailed results for RF and XGBoost in Supplementary Table 4 |
| Application Domains | Organic chemistry, Catalysis, Asymmetric synthesis, Synthetic chemistry, Molecular catalyst design, Drug discovery / medicinal chemistry (indole motifs and atropisomers relevant to pharma) |
80. A database of ultrastable MOFs reassembled from stable fragments with machine learning models, Matter (May 03, 2023)
| Category | Items |
|---|---|
| Datasets | CoRE MOF (ASR subset), Extended CoRE MOF 2019 dataset (unsanitized structures added), MOFSimplify extracted stability dataset (~3,000 MOFs), hMOF, BW-DB, ToBaCCo, ARC-MOF, Ultrastable MOF database (this work) |
| Models | Multi-Layer Perceptron |
| Tasks | Regression, Binary Classification, Feature Extraction |
| Learning Methods | Supervised Learning, Feature Extraction, Feature Selection |
| Performance Highlights | MAE_Td_Celsius: 44, activation_stability_threshold: 0.5, predicted_ultrastable_fraction_ultrastable_MOF_database: 9524of_54139(~18%), predicted_thermally_and_activation_stable_in_new_DB: 25,336of_54,139(~47%), predicted_activation_stable_below_average_thermal: 19,342of_54,139(~36%), predicted_thermally_stable_not_activation_stable: 4,285of_54,139(~8%), ultrastable_counts_hMOF: 416, ultrastable_counts_BW-DB: 767, ultrastable_counts_ToBaCCo: 750, ultrastable_counts_ARC-MOF: 1,564 |
| Application Domains | Materials discovery, Metal-organic frameworks (MOFs) design and screening, Gas storage and separation (methane storage/deliverable capacity), Catalysis (high-temperature catalysis relevance), Mechanical stability assessment of porous materials, In silico database construction for porous materials (including potential application to COFs and ZIFs) |
79. High-throughput printing of combinatorial materials from aerosols, Nature (May 2023)
| Category | Items |
|---|---|
| Datasets | Printed combinatorial gradient films (various materials: Ag/Bi2Te3, Bi2Te2.7Se0.3 with S doping, polyurethane FGP, GO/rGO gradients, etc.), Printing process parameter measurements (ink flow rates, sheath gas flow rates, nozzle sizes, printing speed) |
| Models | None |
| Tasks | Optimization, Hyperparameter Optimization, Data Generation, Experimental Design, Feature Extraction |
| Learning Methods | None |
| Performance Highlights | None |
| Application Domains | Materials science (combinatorial materials discovery), Additive manufacturing / Printed electronics, Thermoelectrics / energy harvesting, Polymer functional grading and mechanics, Nanocomposites and nanomaterials (0D/1D/2D materials), Chemical/materials reaction screening (e.g., GO reduction) |
78. Generative Models as an Emerging Paradigm in the Chemical Sciences, Journal of the American Chemical Society (April 26, 2023)
| Category | Items |
|---|---|
| Datasets | GuacaMol, MOSES (Molecular Sets), Polymer Genome, ANI-2x (ANI2x) |
| Models | Variational Autoencoder, Generative Adversarial Network, Normalizing Flow, Diffusion Model, Graph Neural Network, Recurrent Neural Network, Gaussian Process |
| Tasks | Data Generation, Graph Generation, Sequence-to-Sequence, Optimization, Regression, Language Modeling |
| Learning Methods | Reinforcement Learning, Policy Gradient, Actor-Critic, Deterministic Policy Gradient, Temporal Difference Learning, Adversarial Training, Representation Learning, Active Learning, Supervised Learning |
| Performance Highlights | penalized_logP_benchmark: GraphAF outperformed other common generative models at the time in its ability to generate high penalized logP values (no numeric value provided in text), benchmarking_tasks: MolGAN enabled better predictions on a number of benchmarking tasks (no numeric values provided in text) |
| Application Domains | Chemical sciences, Molecular discovery / drug discovery, Materials science (including organic crystals and functional materials), Polymeric/macromolecular design, Automated/self-driving laboratories / autonomous experimentation, Computational chemistry and molecular simulation (integration with ML interatomic potentials), Synthetic chemistry / retrosynthetic planning |
77. A general-purpose material property data extraction pipeline from large polymer corpora using natural language processing, npj Computational Materials (April 05, 2023)
| Category | Items |
|---|---|
| Datasets | PolymerAbstracts, Materials science corpus (2.4 million abstracts), Polymer-relevant subset of corpus, ChemDNER, Inorganic Synthesis recipes (Materials Science Procedural Text Corpus), Inorganic Abstracts, ChemRxnExtractor, PubMed corpus (referenced), MatBERT pretraining corpus (referenced), ChemBERT pretraining corpus (referenced), Polymer property extraction output (product dataset) |
| Models | BERT, Transformer, Linear Model, Bidirectional LSTM |
| Tasks | Named Entity Recognition, Sequence Labeling, Named Entity Recognition, Structured Prediction, Regression, Information Retrieval |
| Learning Methods | Self-Supervised Learning, Unsupervised Learning, Transfer Learning, Fine-Tuning, Supervised Learning, Weakly Supervised Learning |
| Performance Highlights | Precision (%): 62.5, Recall (%): 70.6, F1 (%): 66.4, ChemDNER F1 (%): 69.2, Inorganic Synthesis recipes F1 (%): 68.6, Inorganic Abstracts F1 (%): 86.0, ChemRxnExtractor F1 (%): 71.4, PolymerAbstracts F1 (%): 65.8, PolymerAbstracts F1 (%): 65.2, PolymerAbstracts F1 (%): 62.6, PolymerAbstracts F1 (%): 57.0, PolymerAbstracts F1 (%): 56.2, Extracted property records (count): 300000, Source abstracts processed: 130000, Processing time: 60 hours on a single Quadro 16 GB GPU |
| Application Domains | Polymers, Materials Science (general), Polymer Solar Cells (organic photovoltaics), Fuel Cells (polymer electrolyte membranes), Supercapacitors (energy storage), Chemical / Organic Chemistry (NER benchmarking datasets) |
76. Evolutionary-scale prediction of atomic-level protein structure with a language model, Science (March 17, 2023)
| Category | Items |
|---|---|
| Datasets | UniRef90 / UniRef50 (training clusters), MGnify90 (MGnify database), Protein Data Bank (PDB), AlphaFold2-predicted structures (augmentation), CAMEO test set, CASP14 test set, Held-out UniRef50 clusters for perplexity evaluation |
| Models | Transformer, Attention Mechanism, Multi-Head Attention |
| Tasks | Language Modeling, Structured Prediction, Representation Learning, Supervised Learning |
| Learning Methods | Self-Supervised Learning, Pre-training, Supervised Learning, Transfer Learning, Fine-Tuning, Knowledge Distillation, Representation Learning |
| Performance Highlights | perplexity_8M: 10.45, perplexity_15B: 6.37, perplexity_3B_on_CAMEO: 5.7, TM-score_15B_on_CAMEO_projection: 0.72, TM-score_15B_on_CASP14projection: 0.55, correlation_perplexity_vs_CASP14_TMscore: -0.99, correlation_perplexity_vs_CAMEO_TMscore: -1.00, correlation_contactprecision_vs_CASP14_TMscore: 0.96, correlation_contactprecision_vs_CAMEO_TMscore: 0.99, ESMFold_TM-score_CAMEO_avg: 0.83, ESMFold_TM-score_CASP14_avg: 0.68, AlphaFold2_TM-score_CAMEO_avg: 0.88, AlphaFold2_TM-score_CASP14_avg: 0.85, ESMFold_LDDT_high_confidence_pLDDT_gt_0.7_on_CAMEO: 0.83, AlphaFold2_LDDT_on_CAMEO: 0.85, median_all-atom_RMSD95_CAMEO: 1.91 Å, median_backbone_RMSD95_CAMEO: 1.33 Å, median_all-atom_RMSD95_pLDDT_gt_0.9: 1.42 Å, median_backbone_RMSD95_pLDDT_gt_0.9: 0.94 Å, DockQ_examples: Glucosamine-6-phosphate deaminase (7LQM) DockQ 0.91; L-asparaginase (7QYM) DockQ 0.97, DockQ_multimer_agreement_with_AlphaFoldMultimer: qualitative DockQ categorization same for 53.2% of chain pairs (on 2,978 complexes), speed_384_residues_on_V100: 14.2 s (≈6× faster than single AlphaFold2 model), speedup_on_short_sequences: up to ~60×, metagenomic_scale_predictions: >617 million sequences folded; ~365 million predictions mean pLDDT>0.5 & pTM>0.5; ~225 million predictions mean pLDDT>0.7 & pTM>0.7, calibration_correlation_pLDDT_vs_AlphaFold_LDDT_on~4000_MGnify_subset: Pearson r = 0.79 |
| Application Domains | Structural biology, Proteomics, Metagenomics, Bioinformatics / Computational biology, Protein design, Biotechnology / drug discovery (potential downstream applications) |
75. AlphaFlow: autonomous discovery and optimization of multi-step chemistry using a self-driven fluidic lab guided by reinforcement learning, Nature Communications (March 14, 2023)
| Category | Items |
|---|---|
| Datasets | AlphaFlow cALD sequence selection campaign data, AlphaFlow reagent volume and reaction time optimization data (volume-time campaign), AlphaFlow digital twin training dataset (480 nm QD campaign), AlphaFlow repository (source data & code) |
| Models | Multi-Layer Perceptron, Gradient Boosting Tree, Multi-Layer Perceptron, Gradient Boosting Tree |
| Tasks | Optimization, Experimental Design, Decision Making, Planning, Control, Binary Classification, Experimental Design |
| Learning Methods | Reinforcement Learning, Model-Based Learning, Supervised Learning, Ensemble Learning |
| Performance Highlights | lambda_AP_shift_vs_conventional_after_6_cycles_nm: 26, photoluminescence_intensity_vs_conventional_percent: 450, RPV_increase_by_4th_cycle_percent: 40, RL_viable_set_found_after_experiments: 4 (87% of known optimum), RL_final_reward_after_100_experiments_percent_of_optimum: 94, ENN_BO_viable_set_found_after_100_experiments: 0 (failed to identify 20-consecutive viable injections), basin_hopping_simulations_needed_for_optimum_function_evals: >50,000 simulated experiments, RL_simulations_experiments_to_reach_94_percent_optimum: 100 real experiments (digital-twin RL campaigns) |
| Application Domains | Chemistry, Materials Science, Nanoscience, Colloidal quantum dot synthesis (CdSe/CdS core-shell QDs), Automated experimentation / Self-driving laboratories, Flow chemistry / microfluidic reaction systems |
74. Adaptively driven X-ray diffraction guided by machine learning for autonomous phase identification, npj Computational Materials (March 02, 2023)
| Category | Items |
|---|---|
| Datasets | Simulated XRD patterns (Li-La-Zr-O and Li-Ti-P-O), ICSD-derived phase lists used for training, Experimental two-phase physical mixtures, In situ LLZO synthesis XRD scans |
| Models | Convolutional Neural Network, Autoencoder |
| Tasks | Multi-label Classification, Classification, Sequence Labeling |
| Learning Methods | Supervised Learning, Ensemble Learning, Active Learning, Pre-training, Unsupervised Learning, Reinforcement Learning |
| Performance Highlights | ensemble F1-score (Li-La-Zr-O, max, 2θ_max=140°): 0.98, ensemble F1-score (Li-Ti-P-O, max, 2θ_max=140°): 0.95, individual prediction max F1-score (Li-* spaces, 2θ_max=120°): 0.91, adaptive sampling F1-score >= 0.88 achieved in effective scan time: 10–15 min, conventional sampling to reach F1 >= 0.88: 25–30 min, minority phase detection rate (adaptive) at >=6 wt%: >=75%, minority phase detection rate (conventional) to reach 75% detection: ≈15 wt%, average scan time (adaptive measurements, experimental mixtures): ≈6 min per pattern, conventional scan time (comparison): 10 min per pattern, detection of short-lived intermediate LaOOH (adaptive): Detected, detection of short-lived intermediate LaOOH (fast 1 min scans): Missed, detection of short-lived intermediate LaOOH (slow 10 min scans): Missed, adaptive scan time when detecting LaOOH: ~4 min |
| Application Domains | Materials science, X-ray diffraction / Crystallography, Solid-state synthesis monitoring, Battery materials characterization (Li-containing chemical spaces), Autonomous / adaptive experimentation and instrumentation control, Spectroscopy and microscopy (generalizable to other modalities) |
73. A multi-modal pre-training transformer for universal transfer learning in metal–organic frameworks, Nature Machine Intelligence (March 2023)
| Category | Items |
|---|---|
| Datasets | 1 million hypothetical MOFs (hMOFs), 20,000 hypothetical MOFs (hMOFs) fine-tuning set, QMOF database (version 13), CoRE MOF (CoREMOF) datasets, Text-mined stability datasets, Pre-training splits (internal) |
| Models | Transformer, BERT, Vision Transformer, Graph Convolutional Network, Graph Neural Network, Multi-Layer Perceptron |
| Tasks | Pre-training tasks (group), Multi-class Classification, Regression, Binary Classification, Regression, Classification, Node Classification |
| Learning Methods | Pre-training, Fine-Tuning, Transfer Learning, Supervised Learning, Multi-Task / Multi-Modal Learning, Attention Mechanism, End-to-End Learning |
| Performance Highlights | accuracy: 0.97, MAE: 0.01, accuracy: 0.98, R2: 0.78, R2: 0.83, R2: 0.77, R2: 0.78, MAE: 0.3, accuracy: 0.76, R2: 0.44, MAE (temperature): 45 °C, relative_performance: outperforms baseline models (energy histogram, descriptor-based ML, CGCNN) across dataset sizes (5k–20k) as shown in Fig. 3c |
| Application Domains | Metal–organic frameworks (MOFs) / porous materials, Materials discovery and design, Gas adsorption and storage (e.g., H2, N2, O2, CO2), Molecular diffusion in porous media, Electronic property prediction (band gap, DFT properties), Materials stability prediction (solvent removal stability, thermal stability), Text-mined experimental property prediction |
72. Accelerating the design of compositionally complex materials via physics-informed artificial intelligence, Nature Computational Science (March 2023)
| Category | Items |
|---|---|
| Datasets | six million texts (literature corpus mined by Pei et al.), NOMAD laboratory / shared materials data, thermodynamic and kinetic databases (CALPHAD and related databases), combinatorial high-throughput experimental materials libraries, ab initio simulation datasets (DFT calculations), Materials literature (heterogeneous corpus) |
| Models | Multi-Layer Perceptron, Graph Neural Network, Random Forest, Attention Mechanism |
| Tasks | Regression, Classification, Image Classification, Semantic Segmentation, Surrogate modeling (mapped to Regression / Model reduction), Information Retrieval, Outlier Detection, Clustering, Dimensionality Reduction, Active Learning (task framed as data acquisition for model improvement) |
| Learning Methods | Supervised Learning, Active Learning, Reinforcement Learning, Semi-Supervised Learning, Unsupervised Learning, Transfer Learning |
| Performance Highlights | discovered_candidates_count: 70 |
| Application Domains | Computational materials science, Alloy design (high-entropy alloys, Invar alloys, superalloys), Ceramics and high-entropy ceramics, Corrosion and surface protection, Battery materials and catalysis (multi-physics problems), Microstructure evolution and mechanical behavior (phase-field, crystal plasticity), Atomistic/molecular dynamics and interatomic potential development, Materials literature mining and knowledge extraction, Sustainable materials design and life-cycle assessment |
71. Biological research and self-driving labs in deep space supported by artificial intelligence, Nature Machine Intelligence (March 2023)
| Category | Items |
|---|---|
| Datasets | NASA GeneLab, NASA Open Science Data Repository (includes GeneLab and other spaceflight-relevant data), BioSentinel dataset (yeast deep-space CubeSat experiment), Nanopore sequencing data generated aboard the ISS, SPOKE knowledge network embeddings (used with transcriptomic spaceflown mouse data), ECG data from astronaut wearable device (used to train ECG Generator model), Various spaceflight image datasets (retinal/OCT, microscopy, behavioural video) |
| Models | Generative Adversarial Network, Variational Autoencoder, Vision Transformer, Transformer, Convolutional Neural Network, Variational Autoencoder, Generative Adversarial Network |
| Tasks | Instance Segmentation, Image Classification, Pose Estimation, Synthetic Data Generation, Data Augmentation, Feature Extraction, Clustering, Dimensionality Reduction, Time Series Forecasting, Anomaly Detection, Language Modeling, Image Generation |
| Learning Methods | Supervised Learning, Transfer Learning, One-Shot Learning, Few-Shot Learning, Pre-training, Fine-Tuning, Generative Learning, Federated Learning, Continual Learning, Contrastive Learning |
| Performance Highlights | performance_level: human-level performance |
| Application Domains | Space biology, Precision astronaut health / space medicine, Multi-omics and genomics (spaceflight sequencing and analysis), Imaging (ocular/retinal imaging, microscopy, behavioural video), Synthetic biology and automated lab automation (self-driving labs), Robotics and microfluidics for automated experiments, Knowledge graph / biomedical knowledge integration, Edge/onboard computing for space systems |
70. A Materials Acceleration Platform for Organic Laser Discovery, Advanced Materials (February 09, 2023)
| Category | Items |
|---|---|
| Datasets | Organic laser dataset (this work) / organic-laser-data (GitHub) |
| Models | None |
| Tasks | Data Generation, Experimental Design, Optimization, Feature Extraction |
| Learning Methods | None |
| Performance Highlights | None |
| Application Domains | Organic semiconductor lasers (OSLs), Accelerated materials discovery / Materials acceleration platforms, Automated synthesis and autonomous laboratories, Optoelectronics (thin-film devices, ASE/lasing characterization), Computational quantum chemistry for structure–property relationships |
69. Machine Learning-Assisted Synthesis of Two-Dimensional Materials, ACS Applied Materials & Interfaces (January 11, 2023)
| Category | Items |
|---|---|
| Datasets | CVD-grown MoS2 dataset (constructed from historical literature) |
| Models | XGBoost, Support Vector Machine, Naive Bayes, Multi-Layer Perceptron |
| Tasks | Binary Classification, Classification, Feature Selection |
| Learning Methods | Supervised Learning, Boosting, Backpropagation, End-to-End Learning |
| Performance Highlights | accuracy: average prediction accuracy of over 88%, AUROC: 0.91, predicted_probability_example_experiment: 86.8913%, validation_probabilities_table2_P(%): [87.6066, 86.2643, 89.8598, 56.0011, 76.2967, 57.0913], recall: 94.1% (SVM recall reached 94.1%, 2% higher than XGBoost for recall) |
| Application Domains | Materials synthesis (chemical vapor deposition), Two-dimensional materials (MoS2), Materials science for electronics and optoelectronics, Accelerated materials discovery and experimental optimization |
68. Toward the design of ultrahigh-entropy alloys via mining six million texts, Nature Communications (January 04, 2023)
| Category | Items |
|---|---|
| Datasets | 6.4 million abstracts (training corpora), 2.6 million candidate alloys (combinatorial search space), Shortlist of 494 HEA candidates, Alloy DOI list used to generate training corpora |
| Models | Multi-Layer Perceptron, Feedforward Neural Network, Principal Component Analysis (PCA) |
| Tasks | Language Modeling, Embedding Learning, Feature Extraction, Dimensionality Reduction, Ranking, Information Retrieval, Named Entity Recognition, Recommendation |
| Learning Methods | Unsupervised Learning, Transfer Learning, Embedding Learning, Representation Learning, Pre-training |
| Performance Highlights | vector_dimension: 200, window_size: 8, training_epochs: 30, training_corpus_size: 6.4M abstracts, selected_candidates_from_search_space: 494 out of 2,600,000, Senkov_alloy_ranking: continually ranked among the top three for 5-component BCC candidates across yearly models, Cantor_alloy_ranking: ranked as the second most promising solid-solution HEA by our method already before 2004, correlation_with_thermodynamic_gamma: positive linear correlation (numeric coefficient not provided), visual_grouping: elements with similar chemical features are grouped together in PCA projection, alloy_name_standardization_effect: all permutations of element order yield the same alloy node in alloyKG (alphabetized), enabling reliable retrieval (qualitative), recommended_candidates: top-ranked alloys such as TiCrFeCoNi (six-component) and TiCrFeCoNiCuZn (seven-component); 494 total shortlisted |
| Application Domains | Materials Science, Metallurgy, High-Entropy Alloys (HEAs) design, Integrated Computational Materials Engineering (ICME), Scientific literature mining / Text mining, Knowledge graph-based literature retrieval |
67. On scientific understanding with artificial intelligence, Nature Reviews Physics (December 2022)
| Category | Items |
|---|---|
| Datasets | 1.6 million molecule search space (Gómez-Bombarelli et al.), SARS-CoV-2 molecular dynamics simulation data, Glycoblocks molecular dynamics data, Large Hadron Collider (LHC) experimental data (ATLAS, CMS etc.), Materials science literature corpus (used for unsupervised word embeddings), Solar System observational data (last ~30 years), Protein structure data / human proteome (AlphaFold outputs and training data), DrugBank chemical space (used in VR exploration), General scientific literature corpora / semantic knowledge networks |
| Models | Graph Neural Network, Transformer, BERT, GPT, Siamese Network, Gradient Boosting Tree, Decision Tree, Random Forest, Recurrent Neural Network, Variational Autoencoder, Feedforward Neural Network, Ensemble Learning, Graph Attention / Graph-based neural approaches (implicit) |
| Tasks | Anomaly Detection, Outlier Detection, Regression, Time Series Forecasting, Representation Learning, Feature Extraction, Information Retrieval, Experimental Design, Clustering, Node Classification, Language Modeling |
| Learning Methods | Unsupervised Learning, Supervised Learning, Weakly Supervised Learning, Reinforcement Learning, Representation Learning, Boosting, Pre-training, Ensemble Learning |
| Performance Highlights | qualitative: high-quality prediction of object’s motion; simultaneously predicts masses correctly, qualitative: BERT/GPT-3 cited as able to help extract scientific knowledge and enable advanced queries in natural-language interaction |
| Application Domains | Physics (theoretical and experimental), Chemistry (molecular design, materials, quantum chemistry), Biology / Structural Biology (protein folding, SARS-CoV-2 spike protein), Quantum Optics / Quantum Computing, High-Energy / Particle Physics (LHC anomaly detection), Astronomy / Astrophysics, Materials Science (literature mining, materials discovery), Mathematics (conjecture generation, theorem guidance), Laboratory Automation and Robotics (automated experiments and discovery) |
64. Into the Unknown: How Computation Can Help Explore Uncharted Material Space, Journal of the American Chemical Society (October 19, 2022)
| Category | Items |
|---|---|
| Datasets | Materials Project, NOMAD, MPDS (Materials Platform for Data Science), NREL MatDB (Computational Science Center - Materials Database), HOIP combinatorial dataset (Lu et al.), Zeolite synthesis dataset (Jensen et al.), Generated reaction dataset (Tempke & Musho / VAE example), Robot photocatalyst experiment dataset (Burger et al.), Dataset of porous/rigid amorphous materials |
| Models | Variational Autoencoder, Generative Adversarial Network, Recurrent Neural Network, Feedforward Neural Network, Machine Learning ForceFields |
| Tasks | Regression, Sequence-to-Sequence, Synthetic Data Generation, Graph Generation, Optimization, Experimental Design, Data Generation |
| Learning Methods | Supervised Learning, Unsupervised Learning, Transfer Learning, Reinforcement Learning, Active Learning |
| Performance Highlights | generated_samples_count: 7,000,000, robot_experiments: 688 experiments, improvement_factor: 6x more active formulations |
| Application Domains | materials discovery, inorganic materials (including perovskites, cathode materials), organic materials and porous organic cages, zeolites / porous crystalline materials, polymeric / amorphous materials (membranes), photocatalysis (hydrogen production), high-throughput experimental robotics / autonomous experimentation, synthesis route prediction / retrosynthesis |
63. The endless search for better alloys, Science (October 07, 2022)
| Category | Items |
|---|---|
| Datasets | published data of ~700 alloys, DFT-calculated physical properties, thermodynamic databases, experimental feedback dataset (measured properties from synthesized candidates), generated candidate compositions (top 1000 candidates), search space of compositions |
| Models | Multi-Layer Perceptron |
| Tasks | Regression, Optimization, Experimental Design, Feature Selection |
| Learning Methods | Supervised Learning, Active Learning |
| Performance Highlights | training_dataset_size: ~700 alloys, discovered_invar_heas_count: 17, lowest_thermal_expansion_coefficient_found: ~2.3e-6 K^-1, prior_HEA_record_thermal_expansion_coefficient: ~1e-5 K^-1, search_space_size: millions of compositions, candidate_pool_size_for_selection: 1000 (generated candidates), iteration_count: 6 cycles, synthesized_candidates_per_cycle: top 3 candidates synthesized, final_discoveries: 17 Invar HEAs discovered |
| Application Domains | Materials Science, Alloy design / High-Entropy Alloys (HEAs), Computational materials discovery, Experimental materials synthesis and characterization |
62. An artificial intelligence enabled chemical synthesis robot for exploration and optimization of nanomaterials, Science Advances (October 07, 2022)
| Category | Items |
|---|---|
| Datasets | Simulated chemical space (extinction spectrum simulation), Experimental UV-Vis spectral datasets from three hierarchically linked chemical spaces, Experimental TEM image dataset (secondary characterization), Multistep synthesis experimental repeats dataset (directed graph multistep runs) |
| Models | None |
| Tasks | Optimization, Experimental Design, Data Generation, Hyperparameter Optimization, Search / Discovery (Exploration) |
| Learning Methods | Evolutionary Learning |
| Performance Highlights | discovery_efficiency_vs_random_search: exploration algorithm found samples belonging to all classes after 78 steps while random search did not find all classes after 200 steps (16 repeats), average_fitness_reached: average fitness of the highest performance samples from different classes eventually reaches 98% of the estimated maximum (simulated benchmark), nanorod_yield_before_optimization: ca. 57%, nanorod_yield_after_optimization: ca. 95%, optimization_runs: 5 steps (115 reactions) per optimization campaign, multisolution_finding: multiple synthetic conditions found corresponding to different morphologies with high spectral similarity (e.g., octahedral, concave octahedral, smooth polyhedral), target_generation: used to create target spectra from 3D nanostructures derived from electron micrographs; enabled optimization toward targets not directly found in exploration, simulation_implementation: GPU-accelerated discrete-dipole approximation implemented in PyDScat-GPU (TensorFlow 2) |
| Application Domains | Nanomaterials discovery, Chemical synthesis automation, Materials science, Spectroscopic characterization (UV-Vis), Robotics for laboratory automation, Computational simulation of optical spectra (discrete-dipole approximation) |
61. Machine learning–enabled high-entropy alloy discovery, Science (October 07, 2022)
| Category | Items |
|---|---|
| Datasets | Invar database (benchmark dataset of Invar alloys), Experimental dataset produced in this work |
| Models | Autoencoder, Gaussian Mixture Model, Gaussian Mixture Model, Multi-Layer Perceptron, Gradient Boosting Tree, Ensemble Learning, Gaussian Process |
| Tasks | Data Generation, Regression, Experimental Design, Optimization, Representation Learning, Feature Extraction |
| Learning Methods | Active Learning, Unsupervised Learning, Supervised Learning, Representation Learning, Ensemble Learning, Feature Extraction |
| Performance Highlights | testing_error_without_physics: 0.19, testing_error_with_physics: 0.14, MAPE_initial_to_three_iterations_FeNiCoCr: 1.5 -> 0.2, discovery_rate_vs_trial_and_error: 5x (fivefold higher discovery rate than trial-and-error, table S2), experimental_alloys_validated: 17 new alloys measured; 2 alloys discovered with TEC ~2 × 10^-6/K at 300 K, representative_experimental_TEC_values: A3 experimental TEC = 1.41 × 10^-6/K; A9 = 2.02 × 10^-6/K; B2 = 4.38 × 10^-6/K; B4 = 4.94 × 10^-6/K |
| Application Domains | metallurgy, materials science, high-entropy alloy discovery, alloy design (Invar and Kovar alloys), computational materials discovery / AI for materials |
60. Autonomous optimization of non-aqueous Li-ion battery electrolytes via robotic experimentation and machine learning coupling, Nature Communications (September 27, 2022)
| Category | Items |
|---|---|
| Datasets | Clio experimental dataset (autonomous electrolyte measurements), AEM-derived simulated dataset / design-space grid, Baseline electrolyte measurements |
| Models | Gaussian Process |
| Tasks | Optimization, Regression, Experimental Design |
| Learning Methods | Supervised Learning, Maximum Likelihood Estimation |
| Performance Highlights | conductivity_optimum_mS_per_cm: 13.7, conductivity_optimum_conditions: EC:DMC 40:60 by mass, 0.9 m LiPF6 (measured at 26–28 °C), number_of_experiments: 42 (live autonomous campaign), mean_repeatability_error_conductivity_percent: ±1.3%, 95%_CI_repeatability_percent: ±3.8%, enhancement_factor_EF_upper_bound_percent: 5, enhancement_factor_EF_trend_over_40_samples_percent: 2.5, acceleration_factor_AF_at_optimum_average: 10x, acceleration_factor_AF_for_98.5%_of_maximum_average: 6x, acceleration_factor_AF_range_at_98.5%: 4.5x–11.5x, overall_time_acceleration_compared_to_random_search: 6x (reported overall acceleration for the work flow), electrolytes_discovered: 6 high-performing electrolytes identified in two work-days, pouch_cell_discharge_capacity_improvement_worst_percent: 5, pouch_cell_discharge_capacity_improvement_best_percent: 13 |
| Application Domains | Battery research / Li-ion electrolytes, Electrochemistry, Materials discovery, Automated experimentation / self-driving laboratory, Energy storage / fast-charging battery development |
59. Data-Driven Materials Innovation and Applications, Advanced Materials (September 08, 2022)
| Category | Items |
|---|---|
| Datasets | Open Quantum Materials Database (OQMD), Materials Project (MP), Automatic-FLOW (AFLOW) / AFLOWLIB, NOMAD (Novel Materials Discovery Laboratory), Computational Materials Repository (CMR) / C2DB, Inorganic Crystal Structure Database (ICSD), Cambridge Structural Database (CSD), Crystallography Open Database (COD), OQMD / MP combined datasets used in case studies, SuperCon database, JARVIS-DFT / JARVIS database, Custom experimental datasets (literature-collected) used across application studies |
| Models | Linear Model, Support Vector Machine, Decision Tree, Random Forest, Gradient Boosting Tree, K-Nearest Neighbor, Gaussian Process, Multi-Layer Perceptron, Convolutional Neural Network, Recurrent Neural Network, Graph Neural Network, Generative Adversarial Network, Variational Autoencoder, Restricted Boltzmann Machine, Ensemble Methods (Bagging/Boosting/AdaBoost), Genetic Algorithm |
| Tasks | Regression, Classification, Binary Classification, Multi-class Classification, Clustering, Dimensionality Reduction, Feature Extraction, Image Classification, Object Detection, Synthetic Data Generation, Graph Generation, Hyperparameter Optimization, Ranking, Data Generation |
| Learning Methods | Supervised Learning, Unsupervised Learning, Deep Learning, Active Learning, Transfer Learning, Bayesian Optimization, Reinforcement Learning, Evolutionary Learning, Ensemble Learning, Active Learning |
| Performance Highlights | RMSE: 0.09 eV, Efrozen_RMSE_train: 0.06 eV, Efrozen_RMSE_test: 0.11 eV, Erelax_train_RMSE: 0.05 eV, Erelax_test_RMSE: 0.10 eV, Pearson_coefficient: 0.79, RMSE_PCE: 1.07% (PCE units), Coverage_calculation_MAE: 0.07 eV, Coverage_calculation_RMSE: 0.10 eV, Coverage_calculation_R2: 0.93, OER_MAE: 0.13 eV, OER_RMSE: 0.18 eV, OER_R2: 0.8, RMSE_bandgap: 0.283 eV, R2: 0.957, R2_bandgap: 0.97, RMSE_bandgap: 0.086 eV, MAE_ΔECO: 0.1 eV, MAD_ΔECO: 0.1 eV, MAE_ΔE_CO_HEA: 0.046 eV, MAE_ΔE_H_HEA: 0.048 eV, RMSE_ΔG_OH: 0.036 eV, R2: 0.993, cn*_MAE: 0.07%, NRR_ΔG_MAE: 0.57 eV, Voltage_MAE: 0.44 V, R2_voltage: 0.86, Elastic_constants_R2_C11: 0.60, R2_C12: 0.79, R2_C44: 0.60, Thermoelectric_S_classification_AUC: 0.96, Thermoelectric_powerfactor_AUC: 0.82, Supercon_classifier_accuracy: 92%, Capacitance_R2: 0.91, Optoelectronics_MAE: 0.086, R2: 0.835, Glass_forming_ability_accuracy: 89%, Dmax_R2: 0.8, Dmax_MAE: 0.21 nm, DeltaTx_MAE: 8.8 K, 2D_PV_classifier_accuracy: 1.0, 2D_PV_precision: 1.0, 2D_PV_recall: 1.0, 2D_PV_AUC: 1.0 |
| Application Domains | Energy conversion (water splitting: HER/OER/OWS), Photovoltaics (perovskite, organic, metal-oxide, 2D PVs), Carbon dioxide reduction reaction (CRR) catalysis, Oxygen reduction reaction (ORR) / fuel cells & metal-air batteries, Nitrogen reduction reaction (NRR) electrocatalysis, Thermoelectric materials discovery, Piezoelectric materials / electrostrains, Rechargeable alkali-ion batteries (electrolytes and electrodes), Supercapacitors, Environmental decontamination (advanced oxidation / PEC processes), Flexible electronics (composite/process optimization), Optoelectronics (2D octahedral oxyhalides), Superconductors (T_c prediction & screening), Metallic glasses (glass-forming ability, Dmax, ΔT_x), Magnetic materials (Curie temperature prediction, permanent/soft magnets), Materials thermodynamic stability and phase prediction, High-throughput computational materials science / databases / workflow automation |
58. Imaging and computing with disorder, Nature Physics (September 2022)
| Category | Items |
|---|---|
| Datasets | DiffuserCam compressive 3D imaging (1.3 megapixel sensor → 100 million voxels), Transmission of natural scene images through a multimode fibre (experimental dataset), Large-scale optical reservoir computing dataset for spatiotemporal chaotic systems prediction, Classification of time-domain waveforms using a speckle-based optical reservoir computer (experimental waveform dataset), Multimode-fibre non-linear classification experiments (dataset used to demonstrate improved classification with nonlinearity), Speckle-based spectrometer spectral-response datasets |
| Models | Perceptron, Convolutional Neural Network, Support Vector Machine, Linear Model, Recurrent Neural Network, Feedforward Neural Network, Graph Neural Network |
| Tasks | Image Classification, Regression, Time Series Forecasting, Object Localization, Image Super-Resolution, Image-to-Image Translation, Classification |
| Learning Methods | Supervised Learning, Backpropagation, End-to-End Learning, Representation Learning |
| Performance Highlights | accuracy_comparison: nonlinear multimode fibre classification accuracy > single-layer neural network accuracy |
| Application Domains | Optics / Photonics / Computational imaging, Acoustics, Radiofrequency communications / Wireless, Seismic waves, Ultrasound imaging, Neuroscience / Functional brain imaging, Quantum information, Neuromorphic computing / hardware |
57. Deep-learning seismology, Science (August 12, 2022)
| Category | Items |
|---|---|
| Datasets | STanford EArthquake Dataset (STEAD), Marmousi2, BP2004, INSTANCE (Italian seismic dataset for machine learning), Authors’ meta-analysis database of published papers, Synthetic and semisynthetic seismic datasets (generated by simulations or GANs), Competition and benchmarking datasets (richterx, AETA, other community benchmarks) |
| Models | Convolutional Neural Network, Recurrent Neural Network, Long Short-Term Memory, Transformer, Attention Mechanism, Autoencoder, Generative Adversarial Network, Capsule Network, U-Net, Graph Neural Network, Physics-Informed Neural Networks, Variational Autoencoder, Multi-Layer Perceptron |
| Tasks | Classification, Binary Classification, Multi-class Classification, Sequence Labeling, Time Series Forecasting, Clustering, Image Denoising, Image Inpainting, Image Super-Resolution, Semantic Segmentation, Object Detection, Regression, Clustering, Node Classification, Optimization, Synthetic Data Generation |
| Learning Methods | Supervised Learning, Unsupervised Learning, Semi-Supervised Learning, Transfer Learning, Reinforcement Learning, Fine-Tuning, Pre-training, Ensemble Learning, Federated Learning, Variational Inference |
| Performance Highlights | speedup: more than 100x, computation_time_reduction: at least an order of magnitude |
| Application Domains | Seismology (earthquake monitoring and seismic imaging), Subsurface characterization (velocity inversion, reservoir properties), Earthquake early warning (EEW) and ground motion prediction, Seismic data denoising and processing (active and passive seismic), Exploratory data analysis of continuous seismic records (e.g., landslides, Mars seismicity, urban noise), Unconventional sensing domains (distributed acoustic sensing, fiber-optic sensing, MEMS accelerometers), Gravitational-wave detection and denoising (LIGO applications), Monitoring moving objects and biosensing (human footsteps, vehicle classification, wildlife detection), Benchmarking and community datasets / AI-for-science infrastructure |
56. Machine learning in the search for new fundamental physics, Nature Reviews Physics (June 2022)
| Category | Items |
|---|---|
| Datasets | ATLAS/CMS LHC collision data (example: 139 fb^-1), LHC trigger data stream (raw collisions), LHCb high-rate data (30 MHz software trigger studies), MicroBooNE data / LArTPC image data, DUNE (simulation and prototyping data), PILArNet, IceCube event data (irregular geometry), LUX dataset (dark matter direct detection), XENON1T dataset, EXO-200 dataset, DarkSide-50 / DarkSide-20k data / prototypes, NEXT / nEXO / PandaX-III (simulated and experimental TPC data), LHC Olympics and Dark Machines challenge datasets, ATLAS/CMS simulated Monte Carlo datasets |
| Models | Multi-Layer Perceptron, Convolutional Neural Network, U-Net, Recurrent Neural Network, Long Short-Term Memory, Graph Neural Network, Autoencoder, Variational Autoencoder, Generative Adversarial Network, Normalizing Flow, Gradient Boosting Tree, XGBoost, Support Vector Machine, Decision Tree, Feedforward Neural Network, Bayesian Network |
| Tasks | Classification, Binary Classification, Multi-class Classification, Regression, Image Classification, Semantic Segmentation, Instance Segmentation, Anomaly Detection, Density Estimation, Data Generation, Clustering, Feature Extraction, Time Series Forecasting, Graph Classification, Node Classification, Image Generation |
| Learning Methods | Supervised Learning, Unsupervised Learning, Semi-Supervised Learning, Weakly Supervised Learning, Self-Supervised Learning, Domain Adaptation, Adversarial Training, Contrastive Learning, Transfer Learning, End-to-End Learning, Representation Learning, Ensemble Learning, Inference-aware Learning / Likelihood-aware Optimization |
| Performance Highlights | light-flavour false-positive rate: 1/390, true positive rate (b-jet efficiency): 70% |
| Application Domains | High-energy particle physics (collider experiments, e.g., LHC: ATLAS, CMS, LHCb), Neutrino experiments (LArTPCs: DUNE, MicroBooNE, SBND, NOvA, KamLAND-Zen, MINERvA, IceCube, KM3NeT/ORCA), Rare event searches (dark matter direct detection, neutrinoless double beta decay: LUX, XENON1T, EXO-200, DarkSide, NEXT, nEXO, PandaX-III, PICO, CRESST), Astrophysics / Cosmology (mentioned as related domains: Gaia, LIGO, Vera C. Rubin Observatory, Square Kilometer Array), Hardware and systems for ML acceleration (FPGAs, GPUs) in scientific triggers and online processing |
55. Enhancing computational fluid dynamics with machine learning, Nature Computational Science (June 2022)
| Category | Items |
|---|---|
| Datasets | two-dimensional Kolmogorov flow, plume configuration, turbulent channel flow at Re_tau = 180, filtered DNS of decaying homogeneous isotropic turbulence, Kraichnan turbulence (2D decaying turbulence), Taylor–Green vortex, reacting and non-reacting finite-volume CFD high-resolution data, particle simulations / PN junction (Poisson solver examples cited), real urban geometry (LES case), flow past a cylinder (canonical ROM example) |
| Models | Convolutional Neural Network, Long Short-Term Memory, Multi-Layer Perceptron, Autoencoder, U-Net, Generative Adversarial Network, Feedforward Neural Network, Random Forest, Gaussian Process, Recurrent Neural Network |
| Tasks | Image Super-Resolution, Regression, Time Series Forecasting, Dimensionality Reduction, Clustering, Optimization, Policy Learning, Feature Extraction, Image-to-Image Translation |
| Learning Methods | Supervised Learning, Reinforcement Learning, Multi-Agent Learning, Transfer Learning, Bayesian Optimization, Unsupervised Learning, Physics-Informed (methodology mentioned: PINNs / physics-informed networks), Representation Learning |
| Performance Highlights | accuracy_vs_Jacobi_solver: outperformed Jacobi solver at low Richardson numbers (Ri), degradation_high_Ri: accuracy degrades at higher Ri, mesh_coarsening_factor: 8-10x coarser in each dimension, agreement: excellent agreement with reference simulations, error: below 10% (for decomposition approach), contextual_note: used as first guess in iterative algorithms, turbulence_statistics: maintained turbulence for an interval long enough to obtain converged turbulence statistics, computational_cost: ≈0.1x (one-tenth) the computational cost compared to reference high-resolution simulation, accuracy_trend: very good agreement with reference high-resolution data initially; errors increase with time, relative_performance: outperforms traditional RANS models based on linear and nonlinear eddy-viscosity models, relative_performance: obtained better results than classical algebraic models, convergence_speed_or_accuracy: GEP outperformed standard LES models in a LES of a Taylor–Green vortex (as reported), generalization: favorable generalization properties across grid sizes and flow conditions (reported qualitatively), accuracy: achieving accurate results when imposing concrete pressure-gradient conditions on a turbulent boundary layer, convergence_speedup: 1.9 to 7.4 times faster convergence than CFD solver alone |
| Application Domains | computational fluid dynamics (CFD), turbulence modeling (RANS and LES), direct numerical simulation (DNS) acceleration, reduced-order modeling (ROM), aerodynamics / aerodynamic optimization, atmospheric boundary layers / weather and climate, combustion (premixed turbulent combustion), multiphase flows (bubble/gas-liquid flows), astrophysics (simulation acceleration use cases), biomedical simulations (PINN applications), high-performance computing (HPC) acceleration |
54. Dataset of solution-based inorganic materials synthesis procedures extracted from the scientific literature, Scientific Data (May 25, 2022)
| Category | Items |
|---|---|
| Datasets | Dataset of solution-based inorganic materials synthesis procedures, Full-text paragraph corpus for BERT pretraining, Paragraph-level labeled dataset for paragraph classification, Annotated materials entity dataset (MER), Solution-based synthesis paragraph corpus (for extraction pipeline) |
| Models | BERT, Bidirectional LSTM, Conditional Random Field, Seq2Seq, Recurrent Neural Network |
| Tasks | Text Classification, Named Entity Recognition, Sequence Labeling, Structured Prediction, Information Retrieval |
| Learning Methods | Self-Supervised Learning, Pre-training, Fine-Tuning, Supervised Learning, Sequence-to-Sequence |
| Performance Highlights | F1: 99.5%, precursors_precision: 0.98, precursors_recall: 0.99, precursors_F1: 0.98, targets_precision: 0.97, Operations_precision: 0.96, Operations_recall: 0.85, Operations_F1: 0.90, Balanced_reactions_precision: 0.94, temperature_F1: 0.94, time_F1: 0.93, atmosphere_F1: 0.94, Quantities_precision: 0.90, Quantities_recall: 0.85, Quantities_F1: 0.87, paragraph_classification_F1: 99.5%, overall_extraction_yield: ~15% (28,749 reactions from 189,553 solution-based paragraphs) |
| Application Domains | Materials science, Inorganic materials synthesis, Scientific text mining / Natural language processing for materials literature, Automated extraction for dataset creation and data-driven synthesis planning |
53. High-entropy nanoparticles: Synthesis-structure-property relationships and data-driven discovery, Science (April 08, 2022)
| Category | Items |
|---|---|
| Datasets | Materials Data Bank (3D atomic coordinates and chemical species), Combinatorial thin-film materials libraries (~342 compositions per batch), Combinatorial HEA nanoparticle microelectrode library (64 cavities), High-throughput computational screening datasets (empirical/CALPHAD/DFT-based), Simulation-generated datasets for ML (first-principles data integrated ML) |
| Models | Gaussian Process, Multi-Layer Perceptron, Graph Neural Network |
| Tasks | Regression, Classification, Optimization, Distribution Estimation, Ranking |
| Learning Methods | Supervised Learning, Active Learning, Reinforcement Learning |
| Performance Highlights | iterations: ~150 iterations |
| Application Domains | Catalysis (thermocatalysis and electrocatalysis), Energy technologies (hydrogen evolution, ammonia synthesis, CO2 reduction, fuel cells), Materials discovery and design (high-entropy alloys, oxides, sulfides, carbides, MXenes), Nanomaterials synthesis and characterization, High-throughput experimentation and closed-loop optimization |
52. Distributed representations of atoms and materials for machine learning, npj Computational Materials (March 18, 2022)
| Category | Items |
|---|---|
| Datasets | Materials Project structures (used to train SkipAtom), Elpasolite formation energy dataset, OQMD (Open Quantum Materials Database) Formation Energy dataset, Matbench test-suite datasets (benchmark tasks), Mat2Vec corpus (materials science literature), Atom2Vec embeddings / dataset (co-occurrence matrix), Processed data & scripts for this study (repository) |
| Models | Feedforward Neural Network, Graph Neural Network, Multi-Layer Perceptron |
| Tasks | Regression, Binary Classification, Feature Extraction |
| Learning Methods | Unsupervised Learning, Supervised Learning, Maximum Likelihood Estimation, Stochastic Gradient Descent, Mini-Batch Learning, Representation Learning, Pre-training |
| Performance Highlights | MAE (eV/atom) - Elpasolite (SkipAtom 30 dim): 0.1183 ± 0.0050, MAE (eV/atom) - Elpasolite (SkipAtom 86 dim): 0.1126 ± 0.0078, MAE (eV/atom) - Elpasolite (SkipAtom 200 dim): 0.1089 ± 0.0061, MAE (eV/atom) - OQMD Formation Energy (Bag-of-Atoms one-hot, sum pooled, 86 dim): 0.0388 ± 0.0002, MAE (eV/atom) - OQMD Formation Energy (Atom2Vec 86, sum): 0.0396 ± 0.0004, MAE (eV/atom) - OQMD Formation Energy (SkipAtom 86, sum): 0.0420 ± 0.0005, MAE (eV/atom) - OQMD Formation Energy (Mat2Vec 200, sum): 0.0401 ± 0.0004, Benchmark summary (qualitative): Pooled Mat2Vec achieved best results in 4 of 8 benchmark tasks; pooled SkipAtom best in 2 of 8. 200-dim representations generally outperform 86-dim. Sum- and mean-pooling outperform max-pooling., Qualitative improvement over existing benchmarks: Authors report outperforming existing benchmarks on tasks where only composition is available (Experimental Band Gap, Bulk Metallic Glass Formation, Experimental Metallicity) — see Fig. 5 for comparisons. |
| Application Domains | Materials science / materials informatics, Computational materials / inorganic crystals, DFT property prediction (formation energy, band gap), High-throughput materials screening, Chemical composition-based property prediction |
50. A self-driving laboratory advances the Pareto front for material properties, Nature Communications (February 22, 2022)
| Category | Items |
|---|---|
| Datasets | Combustion synthesis experiments (Ada self-driving laboratory), Initialization experiments, Spray-coated validation samples, Sputtered palladium reference samples (calibration), XRF hyperspectral maps, Simulated response surface (for benchmarking) |
| Models | Gaussian Process |
| Tasks | Optimization, Regression, Experimental Design, Regression |
| Learning Methods | Supervised Learning, Active Learning |
| Performance Highlights | LOOCV residuals_vs_experimental_uncertainty: comparable (no numeric exact values provided in main text), assigned_uncertainty_for_conductivity_points: 20% of point value (used in heteroskedastic GP), samples_to_outperform_random_search_in_noise_free_scenario: qEHVI required <100 samples to outperform 10,000 random samples, comparison_metrics_used: hypervolume, acceleration factor (AF), enhancement factor (EF), total_physical_experiments_run: 253 experiments, Pareto_front_discovery: four replicate campaigns each generated clear Pareto fronts; specific Pareto-optimal synthesis conditions identified (see main text and Fig. 3) |
| Application Domains | Materials science, Thin-film synthesis and characterization (palladium films), Self-driving laboratories / autonomous experimentation, Materials discovery and optimization (multi-objective), Scalable deposition / spray combustion synthesis, Experimental design and automation |
49. Density of states prediction for materials discovery via contrastive learning from probabilistic embeddings, Nature Communications (February 17, 2022)
| Category | Items |
|---|---|
| Datasets | phDOS dataset (from ref.34, replicated from ref.14), Materials Project eDOS dataset (version 2021-03-22), Materials Project structures without eDOS (candidates), DFT-computed validation set for candidate VB-gap materials |
| Models | Graph Neural Network, Graph Attention Network, Multi-Layer Perceptron, Attention Mechanism, Multi-Head Attention, Variational Autoencoder |
| Tasks | Regression, Distribution Estimation, Binary Classification |
| Learning Methods | Supervised Learning, Contrastive Learning, Variational Inference, Representation Learning, Fine-Tuning, End-to-End Learning |
| Performance Highlights | R2: 0.62, MAE: 0.078, MSE: 0.023, WD: 24, phDOS_CV_MAE: 1.96, phDOS_CV_MSE: 11, phDOS_avgPhonon_MAE: 17.1, phDOS_avgPhonon_MSE: 625, R2: 0.57, MAE: 0.085, MSE: 0.026, WD: 21, phDOS_CV_MAE: 1.32, phDOS_CV_MSE: 10, phDOS_avgPhonon_MAE: 10.6, phDOS_avgPhonon_MSE: 348, R2: 0.63, MAE: 0.086, MSE: 0.029, WD: 33, phDOS_CV_MAE: 3.3, phDOS_CV_MSE: 49, phDOS_avgPhonon_MAE: 26.2, phDOS_avgPhonon_MSE: 2284, R2: 0.57, MAE: 3.8, MSE: 74.5, WD: 0.21, VB_gap_identification_F1: 0.397, VB_gap_identification_Precision: 0.698, VB_gap_identification_Recall: 0.278, VB_gap_discovery_Precision_on_100: 0.47, VB_gap_discovery_Recall_on_100: 1.0, R2: 0.53, MAE: 3.64, MSE: 80.4, WD: 0.27, VB_gap_identification_F1: 0.352, VB_gap_identification_Precision: 0.509, VB_gap_identification_Recall: 0.269, VB_gap_discovery_Precision_on_100: 0.67, VB_gap_discovery_Recall_on_100: 0.27, R2: 0.45, MAE: 0.105, MSE: 0.042, WD: 44, phDOS_CV_MAE: 4.66, phDOS_CV_MSE: 80, phDOS_avgPhonon_MAE: 35.1, phDOS_avgPhonon_MSE: 3392, R2: 0.3, MAE: 4.89, MSE: 120.9, WD: 0.42, VB_gap_identification_F1: 0.182, VB_gap_identification_Precision: 0.263, VB_gap_identification_Recall: 0.139, VB_gap_discovery_Precision_on_100: 0.0, VB_gap_discovery_Recall_on_100: 0.0 |
| Application Domains | Materials science, Computational materials discovery, Spectral property prediction (phonon density of states, electronic density of states), Thermoelectric materials screening, Transparent conductor discovery, High-throughput materials screening |
48. Data-driven modeling and prediction of non-linearizable dynamics via spectral submanifolds, Nature Communications (February 15, 2022)
| Category | Items |
|---|---|
| Datasets | von Kármán beam finite-element simulation, flow past a cylinder (vortex shedding) simulation dataset, sloshing in a water tank (experimental) |
| Models | Linear Model, Polynomial Model |
| Tasks | Dimensionality Reduction, Feature Extraction, Regression, Time Series Forecasting |
| Learning Methods | Supervised Learning, Representation Learning, Feature Learning |
| Performance Highlights | NMTE: 0.027, NMTE_initial_cubic: 1.17 (117%), NMTE_high_order: 0.0386, NMTE: 0.0188, DMD_long_term_behavior: divergent / fails to capture limit cycle |
| Application Domains | structural dynamics / vibration analysis, computational fluid dynamics / flow control, experimental fluid mechanics (sloshing), mechanical engineering (beam models, MEMS), model reduction and dynamical systems analysis |
47. Innovative Materials Science via Machine Learning, Advanced Functional Materials (February 03, 2022)
| Category | Items |
|---|---|
| Datasets | open quantum material database, open inorganic material database, crystallographic open database, thermoelectric open data resource, two-dimensional material database, novel material discovery database, high-throughput combination database of electronic band structure for inorganic scintillator materials, inorganic amorphous database, inorganic crystal structure database (used for XRD patterns), DFT-computational datasets (large DFT-computational data sets and smaller DFT-computed data sets), hypothetical zeolites dataset, small stainless steels dataset (defects and solidification cracking susceptibility) |
| Models | Convolutional Neural Network, Graph Neural Network, Radial Basis Function Network, Multi-Layer Perceptron, Decision Tree, Support Vector Machine, K-Nearest Neighbors, AdaBoost, Naive Bayes, Recurrent Neural Network, Feedforward Neural Network, Graph Convolutional Network, Generative Adversarial Network |
| Tasks | Regression, Classification, Multi-class Classification, Clustering, Dimensionality Reduction, Feature Selection, Optimization, Anomaly Detection, Image Classification, Feature Extraction |
| Learning Methods | Supervised Learning, Unsupervised Learning, Reinforcement Learning, Adversarial Training, Transfer Learning, Pre-training, Ensemble Learning, Few-Shot Learning, Zero-Shot Learning, One-Shot Learning, Transfer Learning |
| Performance Highlights | RMSE_reduction: over 9% |
| Application Domains | materials science, materials discovery, energy materials (e.g., thermoelectrics, battery materials), solid-state materials, catalysis and electrocatalysis, photocatalysis, alloys and metallurgy (stainless steels), polymer materials, semiconductor materials, biocompatible / bio-related materials |
46. An invertible crystallographic representation for general inverse design of inorganic crystals with targeted properties, Matter (January 05, 2022)
| Category | Items |
|---|---|
| Datasets | Materials Project (queried Nov 2019), Ab initio electronic transport database (Ricci et al., reference 1; used via BoltzTraP calculations), Inorganic Crystal Structure Database (ICSD) cross-reference |
| Models | Variational Autoencoder, Convolutional Neural Network, Feedforward Neural Network, Graph Convolutional Network |
| Tasks | Regression, Data Generation, Synthetic Data Generation, Representation Learning, Optimization |
| Learning Methods | Semi-Supervised Learning, Supervised Learning, Representation Learning, Variational Inference, Backpropagation |
| Performance Highlights | case1_Ef=-0.5_validity_rate: 77.8% (14/18), case1_Ef=-0.5_success_rate: 38.9% (7/18), case1_random_success_rate: 10.5% (2,781/26,402), case1_improvement_over_random: 270%, case2_validity_rate: 84.2% (16/19), case2_success_rate: 36.8% (7/19), case2_random_success_rate: 5.5% (3,035/54,925), case2_improvement_over_random: 560%, case3_validity_rate: 42.9% (12/28), case3_success_rate: 7.1% (2/28), case3_random_success_rate: not calculated (lack of complete power factor labels) |
| Application Domains | Inverse design of inorganic crystalline materials, Materials discovery for photovoltaics and optoelectronics (bandgap-targeted design), Thermoelectrics (design for high TE power factor), General materials property-driven design and high-throughput computational materials science |
45. Applied Machine Learning for Developing Next-Generation Functional Materials, Advanced Functional Materials (December 16, 2021)
| Category | Items |
|---|---|
| Datasets | Materials Project, OQMD, AFLOW, ICSD (Inorganic Crystal Structure Database) / experimentally known materials, PbS quantum dots experimental dataset (digitized lab notebooks), Homma ternary electrolyte experimental search, Harada co-doped NASICON-type dataset, Automated perovskite robotics (RAPID) dataset, Organic photovoltaic device dataset (David et al.), Battery cycle dataset (Severson et al.), Electrochemical impedance spectroscopy (EIS) dataset (Dahn et al.), Clustering dataset for solid-state Li-ion conductors (Zhang et al.), Multi-fidelity bandgap datasets (Chen et al.), Semiconducting materials in public databases, Perovskite composition optimization images (well plate images), DFT datasets for catalytic surfaces (Ulissi et al.), MOF generation dataset used with variational autoencoder (Aspuru-Guzik et al.) |
| Models | Graph Convolutional Network, Random Forest, Mask R-CNN, Convolutional Neural Network, Variational Autoencoder, Recurrent Neural Network, Feedforward Neural Network, Gaussian Process, Gaussian Mixture Model, Variational Autoencoder, Generative Adversarial Network |
| Tasks | Regression, Classification, Clustering, Image Classification, Instance Segmentation, Optimization, Clustering, Image Classification, Regression, Anomaly Detection, Data Generation |
| Learning Methods | Supervised Learning, Unsupervised Learning, Reinforcement Learning, Transfer Learning, Few-Shot Learning, Active Learning, Representation Learning |
| Performance Highlights | formation_energy_MAE: 0.039 eV per atom, bandgap_MAE: 0.388 eV, bulk_modulus_MAE: 0.054 log(GPa), EIS_failure_rate: <1% on ~100,000 spectra, DFT_optimizations_needed: as few as 30 DFT optimizations to find most stable polymorphs (per cited study), coverage_RMSE: 0.10 eV, overpotential_RMSE: 0.18 eV, MOF_CO2_capacity: 7.55 mol kg^-1, selectivity_CO2/CH4: 16 (for top generated MOF), initial_dataset_size: ≈3000 materials, reduced_candidates: 82 materials, predicted_high_conductors: 16 with room-temperature conductivity > 1e-4 S cm^-1 (3 with > 1e-2 S cm^-1), candidates_found: 10 candidate double perovskites with OER performance better than LaCoO3 (as reported) |
| Application Domains | Batteries / solid-state electrolytes, Electrocatalysis (OER, CO2 reduction, hydrogen evolution, nitrogen reduction), Optoelectronics (photovoltaics, LEDs, light emitters), Device fabrication and optimization (OLEDs, solar cells), Materials synthesis (nanocrystals, perovskites, MOFs, COFs), High-throughput experimentation and robotics-augmented discovery, Spectroscopy and experimental data analysis (XRD, EIS, NMR, FTIR), Quality control and manufacturing (defect detection in displays), Generative materials design (MOFs, molecules) |
42. Automating crystal-structure phase mapping by combining deep learning with constraint reasoning, Nature Machine Intelligence (September 2021)
| Category | Items |
|---|---|
| Datasets | Multi-MNIST-Sudoku (generated from MNIST & EMNIST), MNIST (single-digit training set), EMNIST (letters A–I used for second Sudoku), Al–Li–Fe oxide synthetic benchmark (phase-mapping dataset with ground truth), Bi–Cu–V oxide experimental dataset, ICDD stick patterns (prototype XRD patterns) |
| Models | ResNet, Multi-Layer Perceptron, Conditional GAN, Gaussian Mixture Model, Gaussian Mixture Model, Capsule Network, ResNet, Non-negative Matrix Factorization |
| Tasks | Image-to-Image Translation, Image Generation, Clustering, Multi-label Classification, Representation Learning, Clustering |
| Learning Methods | Unsupervised Learning, Self-Supervised Learning, Adversarial Training, Pre-training, End-to-End Learning, Stochastic Gradient Descent, Representation Learning, Supervised Learning |
| Performance Highlights | Sudoku accuracy (%): 99 (with 100 unlabelled 9x9 Multi-MNIST-Sudoku instances), digit accuracy: not given numerically in main text but reported to be superior to supervised baselines in Extended Data, reconstruction quality (qualitative): high (DRNets reconstruct mixed images closely; demonstrated in Fig. 1i), Activation accuracy (%) (Al–Li–Fe): 100, Reconstruction loss L1 (Al–Li–Fe): 0.038, Reconstruction loss L2 (Al–Li–Fe): <0.001, Gibbs (%) (Al–Li–Fe): 100, Gibbs-alloy (%) (Al–Li–Fe): 100, Phase field connectivity (%) (Al–Li–Fe): 100, Fidelity loss (Al–Li–Fe): <0.001, Reconstruction loss L1 (Bi–Cu–V): 3.916, Reconstruction loss L2 (Bi–Cu–V): 0.268, Gibbs (%) (Bi–Cu–V): 100, Gibbs-alloy (%) (Bi–Cu–V): 100, Phase field connectivity (%) (Bi–Cu–V): 100, Fidelity loss (Bi–Cu–V): 0.482, Activation accuracy (%) (Al–Li–Fe, k=6): 63.1, Reconstruction loss L1 (Al–Li–Fe, k=6): 29.805, Reconstruction loss L2 (Al–Li–Fe, k=6): 7.169, Gibbs (%) (Al–Li–Fe, k=6): 93.9, Gibbs-alloy (%) (Al–Li–Fe, k=6): 87.0, Phase field connectivity (%) (Al–Li–Fe, k=6): 71.0, Fidelity loss (Al–Li–Fe, k=6): 46.156, relative performance: DRNets substantially outperform supervised CapsuleNet demixing baselines on Multi-MNIST-Sudoku (exact numbers in Extended Data Fig. 4), Sudoku/digit accuracy (qualitative): DRNets better than CapsuleNet supervised baseline |
| Application Domains | materials science (crystal-structure phase mapping, XRD analysis), computer vision / image reasoning (Multi-MNIST-Sudoku demixing), solar fuels / photoelectrocatalysis (experimental follow-up and materials discovery), scientific discovery workflows (integration of prior scientific knowledge and ML) |
41. Accurate prediction of protein structures and interactions using a three-track neural network, Science (August 20, 2021)
| Category | Items |
|---|---|
| Datasets | Protein Data Bank (PDB), CASP14 targets, CAMEO medium and hard targets, Curated set of 693 human protein domains, GPCR benchmark (human GPCRs of currently unknown structure and GPCR sequences with determined structures), Escherichia coli protein complexes (known structures), Cryo-EM map EMD-21645 (IL-12R–IL-12 complex) |
| Models | Transformer, Attention Mechanism, Self-Attention Network, Cross-Attention, Multi-Head Attention, Ensemble Learning |
| Tasks | Sequence-to-Sequence, Regression, Sequence-to-Sequence |
| Learning Methods | End-to-End Learning, Backpropagation, Supervised Learning, Cross-Attention, Stochastic Learning, Ensemble Learning |
| Performance Highlights | qualitative: structure predictions with accuracies approaching those of DeepMind (AlphaFold2) on CASP14 targets, runtime_end_to_end: ~10 min on an RTX2080 GPU for proteins with fewer than 400 residues (after sequence and template search), lDDT_fraction: >33% of 693 modeled human domains have predicted lDDT > 0.8, lDDT_to_Ca-RMSD: predicted lDDT > 0.8 corresponded to an average Cα-RMSD of 2.6 Å on CASP14 targets, TM-score_complexes: many cases with TM-score > 0.8 for two- and three-chain complexes, Cα-RMSD_examples: p101 GBD predicted vs final refined structure: Cα-RMSD = 3.0 Å over the beta-sheets, improved_accuracy: Ensembles and using multiple discontinuous crops generated higher-accuracy models (qualitative improvement reported), CAMEO_benchmark: RoseTTAFold outperformed all other servers on 69 CAMEO medium and hard targets (TM-score values used for ranking), molecular_replacement_success: RoseTTAFold models enabled successful molecular replacement for four challenging crystallographic datasets that had previously eluded MR with PDB models, example_Ca-RMSD_SLP: 95 Cα atoms superimposed within 3 Å yielding a Cα-RMSD of 0.98 Å for SLP C-terminal domain |
| Application Domains | Structural biology, Protein structure prediction, X-ray crystallography (molecular replacement), Cryo-electron microscopy model building/fitting, Protein-protein complex modeling, Functional annotation of proteins / interpretation of disease mutations, Protein design and small-molecule / binder design (computational discovery) |
40. Highly accurate protein structure prediction with AlphaFold, Nature (August 2021)
| Category | Items |
|---|---|
| Datasets | Protein Data Bank (PDB) (training snapshot 28 Aug 2019), CASP14 (Critical Assessment of protein Structure Prediction, CASP14), Recent PDB test set (post-training cutoff), Uniclust30 (v.2018_08), Big Fantastic Database (BFD), UniRef90 (v.2020_01), MGnify clusters (v.2018_12), Distillation predicted-structure dataset (from Uniclust predictions), PDB subsets used in ablations / filtered analyses |
| Models | Transformer, Attention Mechanism, BERT, Graph Neural Network, Multi-Head Attention |
| Tasks | Regression, Feature Extraction, Representation Learning, Classification |
| Learning Methods | Supervised Learning, Self-Supervised Learning, Knowledge Distillation, Self-Training, Fine-Tuning, End-to-End Learning, Ensemble Learning, Representation Learning |
| Performance Highlights | median_backbone_r.m.s.d.95on_CASP14_domains(Cα_r.m.s.d.95): 0.96 Å (95% CI = 0.85–1.16 Å), next_best_method_median_backbone_r.m.s.d.95: 2.8 Å (95% CI = 2.7–4.0 Å), all-atom_r.m.s.d.95_AlphaFold: 1.5 Å (95% CI = 1.2–1.6 Å), all-atom_r.m.s.d.95_best_alternative: 3.5 Å (95% CI = 3.1–4.2 Å), median_backbone_r.m.s.d._recent_PDB_chains: 1.46 Å (95% CI = 1.40–1.56 Å), qualitative_improvement: self-distillation considerably improves accuracy (as shown in ablations and Fig. 4a), distillation_dataset_size: predicted structures for ~355,993 Uniclust sequences used for distillation, ablation_effect_masked_MSA_head: ablations show removing auxiliary masked MSA head reduces performance (see Fig. 4a; exact numeric differences in supplementary material), pLDDT_lDDT-Cα_correlation: least-squares fit lDDT-Cα = 0.997 × pLDDT − 1.17 (Pearson r = 0.76), n = 10,795 chains, domain_GDT_improvements: AlphaFold outperforms alternatives on CASP14 domains (median metrics above); iterative recycling and Evoformer depth contribute to accuracy (see Fig. 4b), Ablation_losses: ablations removing triangle updates/IPA/recycling reduce GDT and lDDT-Cα (Fig. 4a; numeric deltas in Supplementary Methods), inference_time_with_ensembling_256_residues: 4.8 min (single model V100) in CASP14 config; ensembling adds overhead; without ensembling representative timings: 0.6 min for 256 residues, ensemble_vs_single_accuracy: accuracy without ensembling is very close or equal to with ensembling (authors note turning off ensembling for speed), pTM_TM-score_correlation: Least-squares linear fit TM-score = 0.98 × pTM + 0.07 (Pearson’s r = 0.85), n = 10,795 chains |
| Application Domains | structural biology / protein structure prediction, structural bioinformatics, computational biology / proteomics (proteome-scale prediction), structural interpretation for experimental methods (molecular replacement, cryo-EM map interpretation) |
39. Nanoparticle synthesis assisted by machine learning, Nature Reviews Materials (August 2021)
| Category | Items |
|---|---|
| Datasets | CdSe combinatorial synthesis dataset (ref.110), Autonomous microfluidic CdSe quantum dots optimization dataset (SNOBFIT study, ref.55), CsPbBr3 SNOBFIT autonomous platform dataset (ref.56), Lead halide perovskite nanocrystals automated segmented-flow reactor dataset (ref.118), Ensemble neural-network Bayesian optimization dataset for halide exchange (ref.119,120), Silver triangular nanoprism synthesis dataset (ref.137), Literature-assembled metal nanoparticle dataset (illustrative Box 1), Carbon-dot combinatorial dataset (tapioca-flour precursor study), Gold nanocluster literature + new experiments dataset (hybrid NN study), Carbon nanotube growth experimental dataset (autonomous system, ref.158), Web of Science corpus for ‘CdSe nanoparticle synthesis’ (literature survey) |
| Models | Linear Model, Decision Tree, Random Forest, Gradient Boosting Tree, Gaussian Process, Support Vector Machine, Multi-Layer Perceptron, Convolutional Neural Network, Recurrent Neural Network, Feedforward Neural Network |
| Tasks | Regression, Classification, Optimization, Multi-objective Optimization, Active Learning, Reinforcement Learning, Clustering, Dimensionality Reduction |
| Learning Methods | Supervised Learning, Active Learning, Reinforcement Learning, Ensemble Learning, Pre-training, Unsupervised Learning |
| Performance Highlights | Pearson_correlation (wavelength, quantum yield, reaction yield): > 0.9, Pearson_correlation (FWHM): > 0.7, RMSE (predicted thickness of CsPbBr3 nanoplatelets): 0.7556 layers, RMSE (linear model comparison): 1.0266 layers, RMSE (quadratic model comparison): 0.8835 layers, Illustrative_R2 (Box 1 example): 0.75, Scalability_guideline: most suited for datasets < 1,000 samples (training cost scales ~K^3), Optimization_progress (qualitative): Consistent decrease in objective Z with increasing number of experiments; robust model with low error and variance achieved within 20 iterations in some perovskite syntheses, Loss (qualitative): Deep neural network produced significantly lower minimum loss than Bayesian optimization by run 8 (stated as significantly lower; no numeric values provided), RMSE: 0.7556 layers, Pearson_correlation (SPR position prediction): > 0.96 |
| Application Domains | Nanoparticle synthesis, Inorganic semiconductor nanoparticles (quantum dots, perovskite nanocrystals), Metal nanoparticles (gold, silver, palladium, copper, aluminium, nanoclusters), Carbon-based nanoparticles (carbon dots, carbon nanotubes, graphene-based materials), Polymeric nanoparticles (drug delivery, coatings), Materials discovery and optimization, Catalysis, Photovoltaics / optoelectronics, Biomedical imaging / diagnostics, Autonomous laboratories / robotics / microfluidics integration |
38. Physics-informed machine learning, Nature Reviews Physics (June 2021)
| Category | Items |
|---|---|
| Datasets | Harvard Clean Energy Project (HCEP) data set, Tomo-BOS imaging data (espresso cup experiment), 4D-flow MRI (porcine descending aorta) data, Synthetic 3D plasma measurements (electron density and temperature time series), Instrumented indentation data + low-fidelity finite-element simulations, Ab initio molecular simulation data used to train DeePMD, UCI Machine Learning Repository subsets (examples) |
| Models | Multi-Layer Perceptron, Convolutional Neural Network, ResNet, Graph Neural Network, Generative Adversarial Network, Variational Autoencoder, Restricted Boltzmann Machine, Transformer, Gaussian Process |
| Tasks | Regression, Time Series Forecasting, Image Denoising, Image-to-Image Translation, Distribution Estimation, Dimensionality Reduction, Clustering |
| Learning Methods | Supervised Learning, Semi-Supervised Learning, Transfer Learning, Multi-Task Learning, Adversarial Training, Variational Inference, Markov chain Monte Carlo, Active Learning, Gradient Descent, Stochastic Gradient Descent, Variational Inference |
| Performance Highlights | yield_stress_inference_error: reduced from >100% to <5% (reported in ref.64) |
| Application Domains | Fluid dynamics, Biophysics / Biomedical imaging (4D-flow MRI), Plasma physics / Magnetic confinement fusion, Materials science (mechanical property inference, fracture/crack detection), Quantum chemistry / Electronic structure (FermiNet, ab initio), Molecular dynamics (DeePMD; large-scale MD), Geophysics / Seismic inversion, Climate and Earth system science, Turbulence modelling, Computational mechanics / Subsurface flow, Image-based flow inference (experimental flow visualization) |
37. Democratising deep learning for microscopy with ZeroCostDL4Mic, Nature Communications (April 15, 2021)
| Category | Items |
|---|---|
| Datasets | ISBI 2012 Neuronal Segmentation Dataset, EPFL mitochondrial EM dataset, DCIS.COM cells — SiR-DNA nuclear marker (StarDist dataset), YOLOv2 bright-field MDA-MB-231 cell migration dataset, Noise2Void 2D (U-251 paxillin-GFP) dataset, Noise2Void 3D (A2780 lifeact-RFP on cell-derived matrices) dataset, CARE (2D & 3D) actin datasets (LifeAct-RFP and Phalloidin), Deep-STORM raw SMLM datasets (phalloidin-Alexa647 glial cell; DNA-PAINT tubulin U2OS), label-free prediction (fnet) dataset — HeLa TOM20, pix2pix paired dataset (DCIS.COM lifeact-RFP -> nucleus SiR-DNA), CycleGAN datasets (SDC, SRRF, SIM microtubule images), StarDist training dataset (DCIS.COM nuclei masks) |
| Models | U-Net, YOLO, pix2pix, CycleGAN, Generative Adversarial Network |
| Tasks | Image Segmentation, Object Detection, Image Denoising, Image Super-Resolution, Image-to-Image Translation |
| Learning Methods | Supervised Learning, Self-Supervised Learning, Transfer Learning, Pre-training, Fine-Tuning, Adversarial Training |
| Performance Highlights | IoU: 0.90, mAP: 0.60, mSSIM: 0.74, PSNR: 20.4, NRMSE: 0.16, mSSIM: 0.74, PSNR: 24.8, NRMSE: 0.19 |
| Application Domains | Bioimaging / Microscopy, Fluorescence live-cell imaging, Structured Illumination Microscopy (SIM), Spinning-Disk Confocal (SDC), Single-Molecule Localization Microscopy (dSTORM, DNA-PAINT), Electron Microscopy (EM), Cell migration and tracking, Digital pathology / medical imaging (general context mentioned) |
36. Crystallography companion agent for high-throughput materials discovery, Nature Computational Science (April 2021)
| Category | Items |
|---|---|
| Datasets | Synthetic XRD dataset (per phase), BaTiO3 temperature-dependent experimental XRD dataset, ADTA experimental XRD dataset (adamantane-1,3,5,7-tetracarboxylic acid), Ni–Co–Al experimental thin-film library (EDX + XRD), ICSD-derived phase collection for Ni–Co–Al (testing set), CSP-derived ADTA low-energy phases (training/test inputs) |
| Models | Convolutional Neural Network, Feedforward Neural Network |
| Tasks | Classification, Multi-class Classification, Multi-label Classification, Phase Mapping (structured prediction / mapping across composition or temperature), Search / Retrieval (searching predicted phases in CSP database) |
| Learning Methods | Supervised Learning, Ensemble Learning, Stochastic Learning |
| Performance Highlights | accuracy: 0.952, cosine_similarity: 0.941, F1-score: 0.946, matched_classifications: 56/60 (≈0.933), cosine_similarity: 0.735, accuracy: 0.763, F1-score: 0.788, top-3_includes_correct_phase: >90% of classifications contain the correct phase in the top three probabilities, various: Figure 5 compares cosine proximity, accuracy and F1-score across combinations of XCA dataset vs AutoXRD dataset and ensemble vs single-learner models (numerical values reported in figure and text for Ni–Co–Al and ADTA benchmarks) |
| Application Domains | Materials discovery, Crystallography / phase identification from X-ray diffraction (XRD), High-throughput combinatorial materials characterization, Organic polymorph screening, Inorganic alloy phase mapping (Ni–Co–Al), Autonomous experimentation / self-driving laboratories, Potential extension to other 1D characterization modalities (spectroscopy, pair distribution function, XANES, photoelectron spectra, NMR, mass spectra, etc.) |
35. Materials design by synthetic biology, Nature Reviews Materials (April 2021)
| Category | Items |
|---|---|
| Datasets | None |
| Models | None |
| Tasks | Optimization |
| Learning Methods | None |
| Performance Highlights | None |
| Application Domains | Materials synthetic biology, Synthetic biology, Materials science and engineering, Biomedicine, Agriculture, Civil and environmental engineering, Architecture, Product design, Sensing / biosensing, Therapeutics (living therapeutics), Electronics (living electronics / bioelectronics), Energy conversion (microbial fuel cells, biophotovoltaics, artificial photosynthesis), Living building materials / construction, Bioremediation, Biomanufacturing / bioproduction, Biofabrication (3D printing of living materials), Stem- cell differentiation interfaces |
34. Digital Transformation in Materials Science: A Paradigm Change in Material’s Development, Advanced Materials (February 24, 2021)
| Category | Items |
|---|---|
| Datasets | AFLOWlib, Materials Cloud, Materials Genome Initiative (MGI), Metabolomics public repository (example) |
| Models | Convolutional Neural Network, Multi-Layer Perceptron, Graph Neural Network, General ‘learning algorithm’ / ‘AI-software’ (unspecified) |
| Tasks | Regression, Optimization, Experimental Design, Image Classification, Feature Extraction, Ranking, Clustering |
| Learning Methods | Supervised Learning, Active Learning, Transfer Learning, Reinforcement Learning, Representation Learning |
| Performance Highlights | None |
| Application Domains | Materials science (general), Polymer science, Organic synthesis, Catalysis (photocatalysts), Perovskite materials (solar cells), Alloys / metallic glasses, Metal–organic frameworks (MOFs), Battery materials / energy materials, Ceramics, Metabolomics (example for data repositories), Digital fabrication / 3D printing / 4D printing |
33. Bayesian reaction optimization as a tool for chemical synthesis, Nature (February 2021)
| Category | Items |
|---|---|
| Datasets | Suzuki–Miyaura dataset (reaction 1), Buchwald–Hartwig datasets (reactions 2a–2e), Direct arylation / Pd-catalysed C–H functionalization dataset (reaction 3), Mitsunobu reaction dataset (reaction 4), Deoxyfluorination reaction dataset (reaction 5), Quantum mechanical computation outputs for reactions 1–5 (auto-qchem) |
| Models | Gaussian Process, Random Forest, Polynomial Model, Linear Model |
| Tasks | Optimization, Regression, Clustering, Dimensionality Reduction, Feature Selection, Feature Extraction, Active Learning |
| Learning Methods | Supervised Learning, Active Learning, Stochastic Gradient Descent, Maximum Likelihood Estimation, Batch Learning |
| Performance Highlights | worst_case_loss_over_reactions_1_and_2a-e: ≤5% yield (with expected improvement and DFT encodings), standard_deviation_in_outcome_Bayesian_optimization: ≤1.9 (as reported vs DOE methods), statistical_superiority_vs_DOE_mean: p < 0.05 (Bayesian optimization vs DOE designs for mean outcome), reaction_3_success_rate: >99% yield 100% of the time within the experimental budget (for Bayesian optimization runs on reaction 3), human_vs_machine_cross_over: optimizer surpassed human average within 3 batches of five experiments; statistically better after 5 batches (p < 0.05), top_yield_found: 99% yield (identified three distinct conditions giving 99% in 4 rounds of 10 experiments), standard_conditions_benchmark: 60% average yield (replicates: 59% and 60%), top_yield_found: 69% yield (identified by Bayesian optimization in ten rounds of 5 experiments), standard_conditions_benchmark: 36% average yield (replicates: 35% and 36%), relative_performance_vs_GP: Inferior in mean loss, outcome variance and worst-case loss (see Extended Data Table 1), DOE_standard_deviation_range: GSD std ≤6.9; D-optimal std ≤3.3; Bayesian optimization std ≤1.9, DOE_worst_case_loss: GSD worst-case loss ≤16; D-optimal worst-case loss ≤15; Bayesian optimization worst-case loss ≤5 |
| Application Domains | Synthetic organic chemistry, Medicinal chemistry / pharmaceutical development, Computational chemistry (DFT-based feature generation), High-throughput experimentation (HTE) workflows, Automated and human-in-the-loop laboratory optimization |
32. On-the-fly closed-loop materials discovery via Bayesian active learning, Nature Communications (November 24, 2020)
| Category | Items |
|---|---|
| Datasets | Ge–Sb–Te composition spread (177 samples), Fe–Ga–Pd composition spread (benchmark), Raw ellipsometry spectral data (Ge–Sb–Te spread, crystalline & amorphous states), AFLOW.org density functional theory (DFT) computed ternary energy hull (external database), ICSD - Inorganic Crystal Structure Database (external) |
| Models | Markov Random Field, Gaussian Process |
| Tasks | Clustering, Regression, Optimization, Experimental Design, Hyperparameter Optimization |
| Learning Methods | Active Learning, Semi-Supervised Learning |
| Performance Highlights | Fowlkes-Mallows Index (FMI): convergence threshold defined as FMI >= 80%, Iterations to discover optimum (live CAMEO run): 19 iterations (GST467 discovered), Total samples: CAMEO: ~19 iterations to optimum vs full set 177; run time: ~10 h vs ~90 h for full sweep, Iteration lead over GP-UCB (average): approx. 35-iteration lead (CAMEO over GP-UCB) in post-analysis of 100 runs, Ellipsometry prior contribution to lead: 25-iteration lead out of the 35 attributed to ellipsometry prior, Within 1% of optimal in first 20 runs: CAMEO: 31% of runs (over 100) vs GP-UCB: 10% of runs, Not explicitly numeric: GPR implemented via ‘fitrgp’ for propagating functional property predictions; improved predictive accuracy reported qualitatively |
| Application Domains | Materials science, Solid-state materials / phase-change memory (PCM), Photonic switching devices, Autonomous experimentation / robotics for scientific discovery, Synchrotron X-ray diffraction-based characterization, Combinatorial materials discovery |
30. Simple descriptor derived from symbolic regression accelerating the discovery of new perovskite catalysts, Nature Communications (July 14, 2020)
| Category | Items |
|---|---|
| Datasets | Experimental OER dataset of 18 conventional oxide perovskites, Expanded experimental dataset including 5 newly synthesised perovskites (total 23 perovskites), Enumerated candidate perovskite list (screening set) — 3545 oxide perovskites |
| Models | Linear Model |
| Tasks | Regression, Feature Extraction, Ranking, Feature Selection |
| Learning Methods | Supervised Learning |
| Performance Highlights | MAE_eV: 0.021, MAE_meV: 21.0, Pearson_correlation: 0.928, Pareto_front_MAEs_eV: [0.0286, 0.0279, 0.0253, 0.0252, 0.0244, 0.0232, 0.0225, 0.0224, 0.0220] |
| Application Domains | Materials informatics / computational materials discovery, Electrocatalysis (oxygen evolution reaction, OER), Oxide perovskite catalysts design and screening, Experimental materials synthesis and electrochemical characterisation |
29. A mobile robotic chemist, Nature (July 2020)
| Category | Items |
|---|---|
| Datasets | Autonomous robotic search dataset (this work), Initial hole-scavenger screening dataset, Historical photocatalysis dataset (cross-validation), In silico virtual search dataset |
| Models | Gaussian Process, Regression |
| Tasks | Regression, Optimization, Experimental Design, Hyperparameter Optimization |
| Learning Methods | Supervised Learning, Active Learning, Batch Learning, Cross-Validation |
| Performance Highlights | baseline_HER: 3.36 ± 0.30 µmol h⁻¹, best_HER_found_by_search: 21.05 µmol h⁻¹, improvement_factor: ≈6× (21.05 / 3.36), experiments_run: 688 real experiments (43 batches), days_running: 8 days autonomous, best_HER: 21.05 µmol h⁻¹, virtual_searches: 100 virtual searches, avg_virtual_experiments_to_95pct: ≈160 virtual experiments to reach 95% of global maximum HER |
| Application Domains | photocatalysis (hydrogen evolution from water), materials discovery / materials chemistry, autonomous laboratory automation / mobile robotics in chemistry, experimental design and optimization, high-throughput autonomous experimentation |
28. Deep-Learning-Enabled Fast Optical Identification and Characterization of 2D Materials, Advanced Materials (June 09, 2020)
| Category | Items |
|---|---|
| Datasets | Optical microscopy (OM) images of 13 2D materials (training/test split derived from 917 original OM images), CVD graphene OM dataset (for transfer learning experiments), Exfoliated Td-WTe2 OM dataset (for transfer learning experiments), Prediction set: OM images of 17 additional (untrained) 2D materials |
| Models | Convolutional Neural Network, Encoder-Decoder, VGG, U-Net |
| Tasks | Semantic Segmentation, Multi-class Classification, Instance Segmentation, Image Classification, Feature Extraction, Classification |
| Learning Methods | Supervised Learning, Stochastic Gradient Descent, Data Parallel / Ensemble Learning, Transfer Learning, Fine-Tuning |
| Performance Highlights | Global accuracy (by pixel): 0.9689, Mean accuracy (by class): 0.7978, Mean IoU (by pixel): 0.5878, Training time: 30 h 56 min 23 s (on NVIDIA GeForce GTX 1080 Ti, 11 GB), Frames per second (test) CPU: 2.5 fps (224×224 test images), Frames per second (test) GPU: 22.0 fps (224×224 test images), Mean class prediction accuracy (pixel-level, reported elsewhere): 79.78%, Global accuracy (pre-training with 60 images): 0.67 (67%), Comparison (random initialization): Requires at least 240 images to reach comparable accuracy, Mean class accuracy: 0.91 (91%), Quantitative metrics: Not provided as single scalar accuracy in main text; results presented as histograms of projected values and standard deviations showing clear correlations, Description: Ensemble projected values and standard deviations plotted for training set (13 materials) and prediction set (17 materials); qualitative correlation to true bandgap and crystal structure observed |
| Application Domains | 2D materials characterization, Nanomaterials optical imaging, Automated optical microscopy / material searching, Materials property prediction (bandgap, crystal structure) from images, High-throughput experimental screening, Laboratory automation and real-time imaging analysis |
27. Artificial Chemist: An Autonomous Quantum Dot Synthesis Bot, Advanced Materials (June 04, 2020)
| Category | Items |
|---|---|
| Datasets | In-house QD synthesis dataset (flow-synthesized perovskite QD reactions), Pre-training dataset from NNE-UCB optimizations, 150-sample subset used for model comparison |
| Models | Multi-Layer Perceptron, Gaussian Process |
| Tasks | Regression, Optimization, Multi-task Learning, Hyperparameter Optimization |
| Learning Methods | Supervised Learning, Reinforcement Learning, Ensemble Learning, Boosting, Pre-training, Transfer Learning, Multi-Task Learning, Evolutionary Learning |
| Performance Highlights | within_10pct_of_lowest_Z_after_25_experiments: true, pretrained_average_Ep_error_after_25_runs_meV: 1, uninformed_NNE-UCB_average_Ep_error_after_25_runs_meV: 3, pretrained_surpassed_other_methods_for_target_count: 9_of_11, relative_performance_compared_to_NNE: underperformed, within_10pct_of_lowest_Z_after_25_experiments: true |
| Application Domains | Colloidal quantum dot synthesis (metal halide perovskite QDs), Autonomous flow chemistry / microfluidics, Materials discovery and optimization (nanomaterials), Optoelectronic device materials (solar cells, LEDs), Continuous manufacturing / on-demand nanoparticle production |
26. Coevolutionary search for optimal materials in the space of all possible compounds, npj Computational Materials (May 14, 2020)
| Category | Items |
|---|---|
| Datasets | Chemical space of unary and binary compounds constructed from 74 elements (all elements excluding noble gases, rare earth elements, and elements heavier than Pu), Sets of candidate crystal structures per composition (structures generated/optimized with USPEX/VASP) |
| Models | None of the standard ML architectures from the provided list (search uses custom evolutionary / coevolutionary algorithms and physics-based models) |
| Tasks | Optimization, Ranking, Clustering |
| Learning Methods | Evolutionary Learning, Stochastic Learning |
| Performance Highlights | sampled_systems: 600 systems computed in 20 MendS generations (hardness/stability search), search_space_total_binary_systems: 2775 possible binary systems (from 74 elements), best_detected_hardness_diamond_Hv_GPa: 92.7, lonsdaleite_Hv_GPa: 93.6, SiC_Hv_GPa: 33.3, BP_Hv_GPa: 37.2, example_MoB2_Hv_GPa: 28.5, example_MnB4_Hv_GPa (Pnnm ferromagnetic): 40.7, sampled_systems: 450 binary systems over 15 MendS generations (magnetization search), result_top_material: bcc-Fe identified as having the highest zero-temperature magnetization among all possible compounds |
| Application Domains | Computational materials discovery, Theoretical crystallography / crystal structure prediction, Materials design for mechanical properties (hardness, fracture toughness), Magnetic materials discovery (magnetization at zero Kelvin), High-throughput ab initio materials screening |
25. Self-driving laboratory for accelerated discovery of thin-film materials, Science Advances (May 13, 2020)
| Category | Items |
|---|---|
| Datasets | Raw robotic platform data from two optimization runs (Ada thin-film pseudomobility dataset) |
| Models | Phoenics (global Bayesian optimization algorithm), Surrogate model (unspecified) |
| Tasks | Optimization, Experimental Design, Regression, Image Classification |
| Learning Methods | Active Learning, Model-Based Learning, End-to-End Learning |
| Performance Highlights | experiments_per_campaign: 35, campaign_duration: under 30 hours (including restocking consumables) per 35-sample campaign, throughput: one sample synthesized and characterized approximately every 20 min, failure_rate: 1 failed sample per 35-sample campaign, converged_parameters_doping_ratio: ~0.4 equivalents, converged_parameters_annealing_time: ~75 s, image_positions_per_sample: 3 overlapping dark-field images per sample (4000 x 3000 pixels) captured at three locations; spectroscopy and conductance taken at 7 positions |
| Application Domains | Materials Science, Thin-film organic semiconductors (spiro-OMeTAD), Perovskite solar cells, Optoelectronics, Autonomous experimentation / Self-driving laboratories, Clean energy materials discovery |
24. Accelerated discovery of CO2 electrocatalysts using active machine learning, Nature (May 2020)
| Category | Items |
|---|---|
| Datasets | Materials Project-derived copper-containing intermetallics (enumerated surfaces and adsorption sites), Experimental de-alloyed Cu–Al catalyst samples |
| Models | None |
| Tasks | Regression, Dimensionality Reduction, Clustering, Ranking, Optimization |
| Learning Methods | Active Learning, Supervised Learning, Unsupervised Learning |
| Performance Highlights | None |
| Application Domains | electrocatalysis, computational materials discovery / materials science, CO2 electroreduction (CO2-to-C2H4 conversion), high-throughput DFT-driven catalyst screening, experimental electrochemical catalyst testing / chemical energy conversion |
23. Improved protein structure prediction using potentials from deep learning, Nature (January 2020)
| Category | Items |
|---|---|
| Datasets | Protein Data Bank (PDB), CATH (non-redundant domain set), Uniclust30 (2017-10), PSI-BLAST nr dataset (as of 15 December 2017), CASP13 targets / CASP13 dataset |
| Models | Convolutional Neural Network, ResNet, Ensemble (model averaging), Rosetta scoring (Vscore2_smooth and Rosetta relax) |
| Tasks | Distribution Estimation, Binary Classification, Multi-class Classification, Regression, Structured Prediction, Feature Extraction, Data Augmentation |
| Learning Methods | Supervised Learning, Stochastic Gradient Descent, Gradient Descent, Ensemble Learning, Multi-Task Learning, Representation Learning |
| Performance Highlights | distogram–realized-structure correlation (Pearson r): test r = 0.72; CASP13 r = 0.78, DLDDT12 vs lDDT12 correlation: Pearson r = 0.92 (CASP13), effect of downsampling distogram (no distogram): TM score = 0.266 when distance potential removed, contact precision (long-range L,L/2,L/5): AlphaFold contact predictions exceed state-of-the-art (compared to top CASP13 contact methods 498 and 032); exact numbers shown in Fig.1c, CASP13 FM high-accuracy domain counts (TM>=0.7): AlphaFold: 24 out of 43 FM domains; next best: 14 out of 43, Q3 accuracy: 84%, improvement via torsion initialization and torsion potential: Removing torsion potential degrades accuracy slightly (numeric TM drop shown in Fig.4b but small), noisy restarts average TM on test set: 0.641 (noisy restarts) vs 0.636 (sampling from predicted torsions), CASP13 assessor summed z-scores (FM best-of-five): AlphaFold: 52.8; next closest group: 36.6 (group 322), CASP13 combined FM and TBM/FM summed z-score (best-of-five): AlphaFold: 68.3; next closest: 48.2, TM score examples & improvements: AlphaFold often achieves high TM scores; gradient-descent based submissions performed better than fragment-assembly (Extended Data Fig.5b); final Rosetta relax adds +0.007 TM on average, effect of Neff on accuracy (correlation): DLDDT12 vs Neff (normalized by length) Pearson r = 0.634, improved prediction for deeper MSAs: distogram accuracy correlates with Neff (Extended Data Fig.3b) |
| Application Domains | Protein structure prediction / structural biology, Protein fold recognition and homology detection, Protein–protein interface prediction / docking candidates, Ligand binding pocket prediction (structure-guided drug discovery), X-ray crystallography molecular replacement (phasing assistance), General computational biology using MSA-based inference |
21. Data-Driven Materials Science: Status, Challenges, and Perspectives, Advanced Science (September 01, 2019)
| Category | Items |
|---|---|
| Datasets | AFLOW, Materials Project, Open Quantum Materials Database (OQMD), NOMAD (Repository / CoE), HTEM, Organic Materials Database (OMDB), Materials Data Facility (MDF), Crystallography Open Database (COD), Computational Materials Repository (CMR), Materials Cloud / MARVEL NCCR data, SUNCAT / Catalysis Hub, Citrine Informatics (Citrination), Exabyte.io, SpringerMaterials, QCArchive |
| Models | Feedforward Neural Network, Decision Tree, Support Vector Machine, Gaussian Process, Convolutional Neural Network, Autoencoder, Variational Autoencoder, Generative Adversarial Network, Message Passing Neural Network, Graph Neural Network, Gaussian Process |
| Tasks | Regression, Dimensionality Reduction, Clustering, Sequence-to-Sequence, Feature Extraction, Clustering, Optimization, Data Generation, Image Classification |
| Learning Methods | Supervised Learning, Unsupervised Learning, Reinforcement Learning, Evolutionary Learning, Representation Learning, Transfer Learning, Feature Learning |
| Performance Highlights | None |
| Application Domains | Materials science (computational & experimental), Catalysis, Energy materials (photovoltaics, photoelectrochemical water splitting), Organic electronics (OLEDs), Polymers and dielectrics, Alloys and high-entropy alloys, Topological materials / electronic materials, Nanoclusters and surface science, Chemical reaction prediction / cheminformatics |
20. Unsupervised word embeddings capture latent knowledge from materials science literature, Nature (July 2019)
| Category | Items |
|---|---|
| Datasets | Scientific abstracts corpus (collected), DFT thermoelectric power factor dataset (ab initio electronic transport database), Experimental thermoelectric dataset (literature-derived), Elpasolite formation energy dataset, Analogy evaluation sets (created for hyperparameter tuning), English Wikipedia corpus (comparison) |
| Models | Feedforward Neural Network, GloVe, BERT, Multi-Layer Perceptron, Generalized Linear Model |
| Tasks | Unsupervised Learning, Recommendation, Ranking, Binary Classification, Regression, Dimensionality Reduction, Hyperparameter Optimization |
| Learning Methods | Unsupervised Learning, Supervised Learning, Pre-training, Representation Learning, Hyperparameter Optimization |
| Performance Highlights | Spearman_rank_correlation_with_experimental_maximum_power_factor: 59%, Spearman_rank_correlation_with_experimental_maximum_zT: 52%, Average_computed_power_factor_top_10_predictions: 40.8 μW K−2 cm−1, Average_computed_power_factor_candidates: 11.5 μW K−2 cm−1, Average_computed_power_factor_known_thermoelectrics: 17.0 μW K−2 cm−1, Top_10_average_factor_multiple_over_candidates: 3.6x, Top_10_average_factor_multiple_over_known: 2.4x, Percentile_positions_of_top_three_predictions_among_knowns: 99.6th, 96.5th, 95.3rd, DFT_dataset_Spearman_correlation_with_experiment: 31%, MAE_formation_energy_elpasolites: 0.056 eV per atom, accuracy_f1_score_cross_validation: 89% (fivefold cross-validation) |
| Application Domains | Materials science (inorganic materials), Thermoelectrics, Photovoltaics, Topological insulators, Ferroelectrics, Materials discovery / recommendation, Computational materials science (DFT-based property computations) |
19. 2DMatPedia, an open computational database of two-dimensional materials from top-down and bottom-up approaches, Scientific Data (June 12, 2019)
| Category | Items |
|---|---|
| Datasets | 2DMatPedia, Materials Project (MP) subset (input database), JARVIS DFT (used for validation comparison), MPContribs landing page / contributed MP entries |
| Models | None |
| Tasks | Regression, Binary Classification, Clustering, Feature Extraction |
| Learning Methods | None |
| Performance Highlights | None |
| Application Domains | materials discovery, two-dimensional (2D) materials science, optoelectronics (wide-band-gap 2D materials), sensing and catalysis (applications benefiting from high surface-to-volume ratio), spintronics / magnetism (magnetic 2D materials), data-driven materials screening, data mining and machine learning applications |
18. Structure prediction drives materials discovery, Nature Reviews Materials (May 2019)
| Category | Items |
|---|---|
| Datasets | Inorganic Crystal Structure Database (ICSD), Pauling File, Computationally identified layered materials (topology-scaling screening), Ideal nets / topology database (most common ideal nets), High-throughput CSP screening outputs (examples reported) |
| Models | Multi-Layer Perceptron, Feedforward Neural Network, Gaussian Process |
| Tasks | Regression, Optimization, Clustering, Dimensionality Reduction, Feature Extraction, Data Generation |
| Learning Methods | Supervised Learning, Active Learning, Evolutionary Learning, Representation Learning, Multi-Task Learning |
| Performance Highlights | speed_up: 2-4 orders of magnitude (compared with DFT), accuracy: claimed to ‘deliver the same accuracy as first-principles methods’ when trained on sufficient DFT data (qualitative), qualitative: ‘The accuracy of quantum mechanics, without the electrons’ (GAP claim); used to achieve near-DFT accuracy, practical_outcome: Demonstrated acceleration of CSP when combining ML potentials with active-learning loops (reference to Podryabinkin et al.), H3S_Tc_predicted: Tc = 203 K (predicted and experimentally verified), LaH10_experimental_Tc: 250–260 K (experimental reports after CSP prediction), examples_energy_gains: e.g., evolutionary searches found lower-energy structures than data mining for several compounds (differences reported in meV atom−1: 24.7, 5.1, 0.2, 33.3 meV atom−1), organic_semiconductor_mobility: predicted / discovered material with hole mobility 12.3–16.0 cm2 V−1 s−1 (compared with typical <10 cm2 V−1 s−1 for parent molecules) |
| Application Domains | Computational materials discovery, Crystallography / crystal structure prediction (CSP), Superhard materials discovery and mechanical property prediction, Superconductivity (high-Tc conventional superconductors, metal hydrides), Organic semiconductors and organic materials (polymorph prediction, charge mobility), 2D materials and nanoclusters, Surfaces, interfaces and grain boundaries, Battery materials (anode/cathode phases), Photovoltaic materials (bandgap engineering), Catalysis and electride materials (ammonia synthesis, CO2 splitting) |
17. Capturing chemical intuition in synthesis of metal-organic frameworks, Nature Communications (February 01, 2019)
| Category | Items |
|---|---|
| Datasets | HKUST-1 synthesis experiments (robotic platform; reconstructed failed and partially successful experiments) |
| Models | Random Forest, Decision Tree |
| Tasks | Regression, Feature Selection, Optimization, Experimental Design, Dimensionality Reduction |
| Learning Methods | Supervised Learning, Ensemble Learning, Evolutionary Learning |
| Performance Highlights | MAE_cross-validation_percent: <9%, MAE_unseen_data_percent: <14%, variable_importance_example: Temperature has ~3x the impact of reactant ratio (relative importance normalized to 1 for max), best_experimental_BET_m2_per_g: 2045, sampling_efficiency_estimate: 20 intuition-based samples vs ~4-5 thousand samples required without intuition to maintain same sampling accuracy |
| Application Domains | Metal-Organic Framework (MOF) synthesis, Materials science, Chemical synthesis / synthetic inorganic chemistry, Crystallography / crystal growth optimization, Adsorption / surface area optimization, High-throughput robotic experimentation |
16. Active learning for accelerated design of layered materials, npj Computational Materials (December 10, 2018)
| Category | Items |
|---|---|
| Datasets | Three-layer TMDC hetero-structures (H3) — 126 unique structures, Four-layer TMDC hetero-structures (partial set used in BO tests), Adsorption energies dataset (reference dataset used for BO validation) |
| Models | Gaussian Process |
| Tasks | Regression, Optimization, Feature Extraction, Feature Selection |
| Learning Methods | Supervised Learning, Active Learning, Maximum Likelihood Estimation |
| Performance Highlights | training_split_threshold: training sets with fewer than 60% of structures did not produce reliable predictions; >60% showed no additional improvement, evaluation_runs: 100 independent GPR models (randomly selected training sets) used to collect statistics and average out effects from initial training data selection, band_gap_model_training_fraction_used_for_figure: 60% of structures randomly selected for training in shown example, predicted_vs_ground_truth: Figures demonstrate predicted vs ground truth band gap, dispersion curves, and EFF curves with 95% confidence intervals (no single numeric MSE in main text), BO_runs: 500 independent BO runs (different random initial training seeds), max_band_gap_success_rate: 79% of BO runs correctly found the structure with the maximum band gap (1.7 eV); 15% found second-best (1.5 eV); 5% found third-best (1.3 eV), desired_band_gap_1.1eV_success_rate: For searching band gap closest to 1.1 eV, MoSe2-WSe2-WSe2 (band gap 1.05 eV) was returned in 91% of 500 runs, EFF_top_found_rate: In band gap (EFF) optimization, one of the top four (five) optimal structures is found within 30 BO iterations in over 95% of the 500 runs, adsorption_dataset_result: On the adsorption energies dataset, after evaluating only 20% of the dataset, 82% of 500 independent BO runs successfully identified the pair with minimum adsorption energy |
| Application Domains | Materials design and discovery, Two-dimensional materials (transition metal dichalcogenide heterostructures), Optoelectronics (band gap engineering for solar cells; Shockley–Queisser limit relevance), Thermoelectrics (electronic transport component and thermoelectric Electronic Fitness Function), Catalysis / surface science (validation on adsorption energy dataset) |
15. Molecular Dynamics Simulation for All, Neuron (September 19, 2018)
| Category | Items |
|---|---|
| Datasets | Experimental structural datasets (X-ray crystallography, cryo-EM, NMR, EPR, FRET; implied Protein Data Bank structures), Web of Science publication set (top 250 journals) for the term ‘molecular dynamics’, MD simulation trajectories (all-atom and coarse-grained trajectories generated in cited studies and by reviewed work) |
| Models | None |
| Tasks | Regression, Ranking |
| Learning Methods | None |
| Performance Highlights | None |
| Application Domains | Molecular dynamics / Computational biophysics, Structural biology, Molecular and cellular physiology, Neuroscience (proteins relevant to neuronal signaling, ion channels, neurotransmitter transporters, GPCRs), Drug discovery and medicinal chemistry (lead optimization, virtual screening, allosteric modulator design, biased ligand design), Protein folding and aggregation studies |
14. Deep neural networks for accurate predictions of crystal stability, Nature Communications (September 18, 2018)
| Category | Items |
|---|---|
| Datasets | Unmixed garnets dataset, Mixed garnets dataset (computed orderings), Unmixed perovskites dataset, Mixed perovskites dataset (A- and B-site mixing), Extended generated candidate set (garnets) |
| Models | Feedforward Neural Network, Multi-Layer Perceptron, Deep Neural Network |
| Tasks | Regression, Binary Classification |
| Learning Methods | Supervised Learning, Backpropagation, Stochastic Gradient Descent |
| Performance Highlights | RMSE: 12 meV atom−1, MAE_training: 7 meV atom−1, MAE_validation: 10 meV atom−1, MAE_test: 9 meV atom−1, MAE_training: 22 meV atom−1, MAE_validation: 26 meV atom−1, MAE_test: 26 meV atom−1, MAE_training: ≈11–12 meV atom−1, MAE_validation: ≈11–12 meV atom−1, MAE_test: ≈11–12 meV atom−1, std_predicted_Ef_across_orderings: 2.8 meV atom−1, MAE_training: 21 meV atom−1, MAE_validation: 34 meV atom−1, MAE_test: 30 meV atom−1, MAE_range: 22–39 meV atom−1, Garnet_accuracy_at_Ehull_0_meV/atom: >90% (unmixed; C-mixed DNN also >90% for mixed), Perovskite_accuracy_at_Ehull_0_meV/atom: >80%, Perovskite_accuracy_at_Ehull_30_meV/atom: >70% |
| Application Domains | Materials science, Inorganic crystal stability prediction, Garnet materials (C3A2D3O12), Perovskite materials (ABO3), Computational materials discovery / high-throughput screening |
13. Accelerated discovery of stable lead-free hybrid organic-inorganic perovskites via machine learning, Nature Communications (August 24, 2018)
| Category | Items |
|---|---|
| Datasets | 212 selected HOIPs (training/test), 346 HOIPs (initial collected dataset), 5158 unexplored HOIPs (prediction set), 5504 possible HOIPs (combinatorial space) |
| Models | Gradient Boosting Tree, Support Vector Machine, Gaussian Process, Decision Tree, Multi-Layer Perceptron |
| Tasks | Regression, Feature Selection, Feature Extraction, Hyperparameter Optimization |
| Learning Methods | Supervised Learning, Ensemble Learning, Boosting |
| Performance Highlights | R2: 0.970, Pearson_r: 0.985, MSE: 0.086, DFT_agreement: ΔEg ≤ 0.1 eV (for six selected HOIPs) |
| Application Domains | Materials science, Computational materials design, Hybrid organic-inorganic perovskites (HOIPs), Photovoltaics / solar cell materials screening, High-throughput materials screening (ML + DFT combined workflows) |
12. Inverse molecular design using machine learning: Generative models for matter engineering, Science (July 27, 2018)
| Category | Items |
|---|---|
| Datasets | chemical space project, Materials Project, MoleculeNet |
| Models | Variational Autoencoder, Autoencoder, Recurrent Neural Network, Long Short-Term Memory, Attention Mechanism, Generative Adversarial Network, Graph Neural Network, Message Passing Neural Network, Gaussian Process, Convolutional Neural Network |
| Tasks | Regression, Data Generation, Sequence-to-Sequence, Text Generation, Graph Generation, Optimization, Representation Learning |
| Learning Methods | Supervised Learning, Semi-Supervised Learning, Reinforcement Learning, Adversarial Training, Evolutionary Learning, Policy Gradient, Q-Learning, Representation Learning, Backpropagation, Active Learning, Adversarial Training |
| Performance Highlights | None |
| Application Domains | Drug discovery / pharmaceuticals, Organic photovoltaics, Organic redox flow batteries, Organic light-emitting diodes, Catalysis and reaction discovery, Inorganic materials (dielectric and optical materials, photoanodes, battery electrolytes), Automated materials discovery / closed-loop experimental laboratories, Quantum chemistry / property prediction |
11. Insightful classification of crystal structures using deep learning, Nature Communications (July 17, 2018)
| Category | Items |
|---|---|
| Datasets | AFLOWLIB elemental solid database (pristine subset), Defective dataset (generated from pristine AFLOWLIB subset), AFLOW Library of Crystallographic Prototypes (used to generate transition-path structures), Processed two-dimensional diffraction fingerprint images (DF) derived from structures |
| Models | Convolutional Neural Network |
| Tasks | Image Classification, Multi-class Classification, Feature Extraction, Representation Learning |
| Learning Methods | Supervised Learning, Mini-Batch Learning, Backpropagation, Gradient Descent, Representation Learning, Feature Extraction |
| Performance Highlights | accuracy_pristine_train: 100.0%, accuracy_pristine_test: 100.0%, robustness_random_displacement_up_to_sigma_0.06A: 100.0%, robustness_vacancies_up_to_40%: 100.0%, robustness_vacancies_at_60%: >97% (reported), prediction_time_per_image_CPU: ≈70 ms (including reading time) on quad-core Intel i7-3540M, training_time_CPU: ≈80 minutes on quad-core Intel i7-3540M, interpretable_filters: yes (attentive response maps show that learned filters correspond to diffraction peak arrangements / class templates) |
| Application Domains | materials science, computational materials science, crystallography / solid-state physics, high-throughput materials discovery, atom probe tomography (local microstructure determination) |
10. Machine learning for molecular and materials science, Nature (July 2018)
| Category | Items |
|---|---|
| Datasets | Inorganic Crystal Structure Database (ICSD), Elpasolite dataset (ABC2D6), 20,000+ crystalline and non-crystalline compounds training set, AFLOWLIB, Computational Materials Repository (CMR), GDB (databases of hypothetical small organic molecules), Harvard Clean Energy Project dataset, Materials Project, NOMAD, Open Quantum Materials Database (OQMD), NREL Materials Database, TEDesignLab, ZINC, ChEMBL, ChemSpider, Citrination, Crystallography Open Database, CSD (Cambridge Structural Database), MatNavi, MatWeb, NIST Chemistry WebBook, NIST Materials Data Repository, PubChem |
| Models | Naive Bayes, Decision Tree, Random Forest, Support Vector Machine, Multi-Layer Perceptron, Convolutional Neural Network, Generative Adversarial Network, Recurrent Neural Network, Graph Neural Network, Boltzmann Machine, Bayesian Network, Neural Turing Machine, Gaussian Process, Perceptron, Message Passing Neural Network |
| Tasks | Supervised Learning, Unsupervised Learning, Semi-Supervised Learning, Classification, Regression, Sequence-to-Sequence, Image Classification, Data Generation, Active Learning, Hyperparameter Optimization, Representation Learning |
| Learning Methods | Supervised Learning, Unsupervised Learning, Semi-Supervised Learning, Reinforcement Learning, Active Learning, Meta-Learning, One-Shot Learning, Adversarial Training, Representation Learning, Imitation Learning |
| Performance Highlights | new_compounds_verified: 12, accuracy: around 80%, success_rate: 89%, coverage_increase: 6x |
| Application Domains | Molecular chemistry, Materials science, Computational chemistry / electronic structure, Drug discovery / medicinal chemistry, Crystallography / crystal engineering, Surface science and microscopy, High-throughput virtual screening, Experimental design and autonomous experimentation, Text mining / literature extraction |
9. ChemOS: Orchestrating autonomous experimentation, Science Robotics (June 20, 2018)
| Category | Items |
|---|---|
| Datasets | 1100 experiments designed by ChemOS (direct-inject HPLC calibration meta-lab), in-house robot color and cocktail spaces dataset, direct-inject sampling dataset for real-time reaction monitoring, autocalibration / tequila sunrise mixing experiment dataset |
| Models | Gaussian Process, Multi-Layer Perceptron, Perceptron, Other (PHOENICS) |
| Tasks | Experimental Design, Optimization, Online Learning, Experimental Design, Active Learning |
| Learning Methods | Online Learning, Active Learning, Reinforcement Learning |
| Performance Highlights | None |
| Application Domains | Chemistry, Materials Science, Organic Synthesis, Automated / Autonomous Laboratories, Robotics, Analytical Chemistry (HPLC, NMR, reaction monitoring), High-throughput Experimentation, Human-AI Collaborative Experimentation |
8. Accelerated discovery of metallic glasses through iteration of machine learning and high-throughput experiments, Science Advances (April 13, 2018)
| Category | Items |
|---|---|
| Datasets | Landolt-Börnstein (LB) melt-spinning dataset, Landolt-Börnstein (LB) sputtering dataset, HiTp combinatorial sputter co-deposition — Co-V-Zr (this work), HiTp combinatorial sputter co-deposition — Co-Ti-Zr, Co-Fe-Zr, Fe-Ti-Nb (this work), Combinatorial candidate search space (screened computationally) |
| Models | Random Forest |
| Tasks | Binary Classification, Regression, Ranking, Feature Selection, Hyperparameter Optimization |
| Learning Methods | Supervised Learning, Ensemble Learning, Stacking, Active Learning |
| Performance Highlights | AUC_ROC: 0.88, Grouping-test_accuracy_before_PCT: 75.9%, Grouping-test_accuracy_after_PCT: 76.8%, Log-loss_before_vs_after_PCT_for_Co-V-Zr_map: 3.56 versus 1.75, Log-loss_Co-V-Zr: reduced from 1.75 to 0.28, AUC_ROC_over_sputtered_dataset_generations: improvement from 0.66 (first-gen) to 0.80 (second-gen/third-gen combined reference in Fig.5C), Log-loss_Co-Ti-Zr: reduced from 1.58 to 0.39 (first-gen to second-gen), Log-loss_Co-Fe-Zr: reduced from 1.70 to 0.49 (first-gen to second-gen), Log-loss_Fe-Ti-Nb: reduced from 2.37 to 1.48 (first-gen to second-gen), AUC_ROC_progression_first_to_third_generation: AUC increases from 0.66 (first-gen) to 0.80 (third-gen aggregated comparison), Stacked_model_effect: stacked approach shows most improvement for sputtered-synthesis prediction (qualitative ROC improvement described) |
| Application Domains | Materials science, Metallurgy / Metallic glasses, High-throughput experimentation (combinatorial materials synthesis), Materials discovery and design, Computational materials science / data-driven materials screening |
7. Two-dimensional materials from high-throughput computational exfoliation of experimentally known compounds, Nature Nanotechnology (March 2018)
| Category | Items |
|---|---|
| Datasets | Inorganic Crystal Structure Database (ICSD), Crystallographic Open Database (COD), Derived 2D materials database (this work), Materials Project (mentioned) |
| Models | None |
| Tasks | Data Generation, Feature Extraction, Pattern Recognition, Clustering |
| Learning Methods | None |
| Performance Highlights | None |
| Application Domains | Two-dimensional materials discovery, Computational materials science, Electronic and optoelectronic materials, Spintronics and data storage (magnetic 2D materials), Topological materials (quantum spin Hall insulators), High-throughput materials screening and database generation |
6. Accelerated Discovery of Large Electrostrains in BaTiO3-Based Piezoelectrics Using Active Learning, Advanced Materials (January 08, 2018)
| Category | Items |
|---|---|
| Datasets | Initial training set of synthesized BTO-based compounds, Unexplored composition search space, Newly synthesized compounds (iterative active learning outputs) |
| Models | Support Vector Machine, Gradient Boosting Tree, Ensemble Learning |
| Tasks | Regression, Feature Selection, Feature Extraction, Experimental Design, Ranking, Optimization |
| Learning Methods | Supervised Learning, Active Learning, Ensemble Learning, Boosting |
| Performance Highlights | best_model_selection_criterion: least cross-validation error (qualitative), bootstrap_samples_for_uncertainty: 1000, guided_discovery_outcome_bipolar_strain: 0.23% (bipolar electrostrain at 20 kV cm^-1 for discovered compound), guided_discovery_outcome_unipolar_strain: 0.19% (maximum unipolar electrostrain at 20 kV cm^-1 for discovered compound), features_initial: 71, features_after_correlation_pruning: 18, important_features_identified: direction of dependence of C–T (NCT) and T–O (NTO) transition temperatures on dopants (example), bootstrap_samples: 1000, design_strategies_compared: exploitation, exploration, trade-off (efficient global optimization), random, iterations: 5 iterative rounds; 5 compounds predicted and synthesized per strategy (20 compounds total), successful_improvements: 9 of 20 synthesized compounds had larger electrostrains than the best in training set, statistical_significance: Fisher p-value < 0.001, best_discovered_composition_strain: 0.23% bipolar at 20 kV cm^-1 (Ba0.84Ca0.16)(Ti0.90Zr0.07Sn0.03)O3, electrostrictive_coefficient_Q33_for_best: 0.106 m^4 C^-2, design_strategy_best: trade-off between exploration and exploitation (referred to as efficient global optimization) “performs in a superior manner to the others” |
| Application Domains | Accelerated materials discovery, Piezoelectric / electrostrictive materials, Experimental materials synthesis and characterization, Computational materials modeling (DFT, Landau theory, phase-field simulations), Optimal experimental design / active learning in materials science |
5. Virtual screening of inorganic materials synthesis parameters with deep learning, npj Computational Materials (December 01, 2017)
| Category | Items |
|---|---|
| Datasets | Text-mined synthesis database (SrTiO3 non-augmented), Augmented synthesis dataset (SrTiO3 neighborhood), Text-mined synthesis data for BaTiO3, Text-mined synthesis data for TiO2, Text-mined synthesis data for MnO2, Word embedding (materials) training corpus / word2vec vectors |
| Models | Variational Autoencoder, Feedforward Neural Network, Multi-Layer Perceptron, Generalized Linear Model |
| Tasks | Dimensionality Reduction, Binary Classification, Feature Extraction, Data Augmentation, Synthetic Data Generation, Representation Learning |
| Learning Methods | Unsupervised Learning, Supervised Learning, Generative Learning, Representation Learning, Feature Learning |
| Performance Highlights | accuracy_30-D_canonical: 74%, std_30-D_canonical: 3%, accuracy_2-D_PCA: 63%, std_2-D_PCA: 3%, accuracy_10-D_PCA: 68%, std_10-D_PCA: 6%, accuracy_2-D_VAE_features_classifier: 63%, std_2-D_VAE: 3%, accuracy_10-D_VAE_features_classifier: 74%, std_10-D_VAE: 6%, sampling_attempts_for_>=95%_chance_trick_classifier: 5 attempts (to exceed 95% chance of producing at least one virtual sample that tricks the LR classifier), trials: 50, reconstruction_error_with_augmentation: reduced (quantitative values not reported in main text), contextual_performance_note: training and validation cross-entropy losses decreased when using augmented dataset (Fig. 2b) |
| Application Domains | Inorganic materials synthesis, Materials science (perovskites, TiO2 polymorphs, MnO2 polymorphs), Catalysis, Energy storage (battery electrodes), Photocatalysis, Computational materials screening / synthesis planning |
4. Design of efficient molecular organic light-emitting diodes by a high-throughput virtual screening and experimental approach, Nature Materials (October 2016)
| Category | Items |
|---|---|
| Datasets | Virtual chemical library (enumerated candidates), TD-DFT screened subset, TD-DFT-derived training/validation data for ML (empirical model), Calibration dataset (experiment vs theory), Experimental device dataset (synthesized leads) |
| Models | Multi-Layer Perceptron, Linear Model |
| Tasks | Regression, Ranking, Feature Extraction, Hyperparameter Optimization |
| Learning Methods | Supervised Learning, Bayesian Optimization, Backpropagation |
| Performance Highlights | R^2 (vs TD-DFT labels): 0.94, R^2 (vs TD-DFT labels): 0.80, Top-5% hit fraction: varies with training size (plotted in Fig.3a), ΔEST prediction R^2 (Arrhenius activation energy comparison): 0.84, ΔEST RMSE: 0.08 eV, Mean unsigned error (emission wavelength): 7 nm, Mean unsigned error (ΔEST): 0.1 eV, Mean unsigned error (f): 0.05, Mean unsigned error (kTADF): 0.1 μs^-1 |
| Application Domains | Organic electronics, Organic light-emitting diodes (OLEDs), Materials discovery / computational materials design, Cheminformatics / virtual screening, Optoelectronics |
3. Machine-learning-assisted materials discovery using failed experiments, Nature (May 2016)
| Category | Items |
|---|---|
| Datasets | Archived laboratory notebook reactions (dark reactions) - curated dataset, Sampled commercially available diamines (experimental validation set), Candidate diamine pool (eMolecules), Cambridge Structural Database (CSD) check |
| Models | Support Vector Machine, Decision Tree, Random Forest, Generalized Linear Model |
| Tasks | Classification, Binary Classification, Feature Extraction, Feature Selection |
| Learning Methods | Supervised Learning, Ensemble Learning |
| Performance Highlights | test-set_accuracy_all_reaction_types: 78%, test-set_accuracy_vanadium-selenite_only: 79%, average_accuracy_over_15_splits: 74%, experimental_success_rate_for_model_recommendations: 89%, human_intuition_experimental_success_rate: 78%, accuracy_with_only_six_selected_features: 70.7% |
| Application Domains | Materials discovery, Inorganic–organic hybrid materials synthesis, Hydrothermal/solvothermal synthesis, Crystallization prediction / crystal formation, Cheminformatics-driven descriptor generation, Experimental planning / reaction recommendation |
2. Accelerated search for materials with targeted properties by adaptive design, Nature Communications (April 15, 2016)
| Category | Items |
|---|---|
| Datasets | Initial training set of 22 alloys, Search space of potential alloys, Synthesized predicted alloys (design loop outputs), Feature descriptors (per-alloy feature vectors) |
| Models | Gaussian Process, Support Vector Machine |
| Tasks | Regression, Optimization, Experimental Design, Feature Selection |
| Learning Methods | Supervised Learning, Active Learning |
| Performance Highlights | best_discovered_ΔT_K: 1.84, synthesized_candidates: 36, improved_alloys_ΔT<3.15K: 14, training_set_size: 22, search_space_size: 797,504, bootstrap_samples_for_uncertainty: 1000, Mann-Whitney_U: 172, Mann-Whitney_z(sd): 3.6, Mann-Whitney_p: <0.001, probability_random_occurrence: 3.7e-4, relative_performance_samples_4_to_8: nearly identical to SVR rbf:KG, relative_performance_samples_2_and_3: best among combinations for sample sizes 2 and 3 |
| Application Domains | Materials discovery, Materials science (shape memory alloys, NiTi-based SMAs), Adaptive experimental design / closed-loop experimentation, Computational materials design / surrogate modeling |
1. The Open Quantum Materials Database (OQMD): assessing the accuracy of DFT formation energies, npj Computational Materials (December 11, 2015)
| Category | Items |
|---|---|
| Datasets | Open Quantum Materials Database (OQMD), Inorganic Crystal Structure Database (ICSD) (structures used as inputs), SGTE Solid SUBstance (SSUB) database, Thermodynamic database at the Thermal Processing Technology Center (IIT), Combined experimental formation-energy comparisons (deduplicated), Actinide thermodynamics review (actinide oxides), Materials Project (comparison set) |
| Models | None |
| Tasks | Regression, Binary Classification, Synthetic Data Generation, Optimization |
| Learning Methods | None |
| Performance Highlights | None |
| Application Domains | Computational materials science, Thermochemistry and phase stability analysis, Crystallography / crystal-structure databases, Materials discovery (prediction of new compounds), Battery and energy materials applications (examples/use cases referenced) |